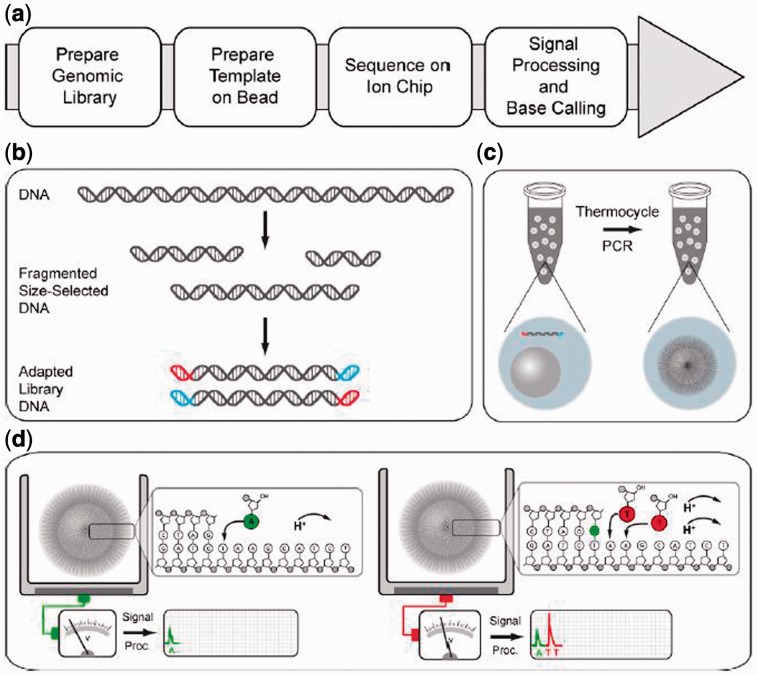
Fig. 1.
Ion sequencing work flow. The overall workflow is shown in (a). A genome library is prepared by fragmenting and size-selecting DNA, followed by the ligation of forward and reverse adapters (b). Each adapter-ligated template is clonally amplified onto a bead, so that each bead contains many copies of the same DNA template (c). Sequence on the chip, sequencing primers and DNA polymerase are then bound to the beads, which are pipetted into wells on the chip (d). The chip is then repeatedly flooded by nucleotides, which, when binding to the complementary nucleotide on a template, release an ion. At each flow, the electrical signal at each well is measured, indicating the number of incorporations [Figure adapted from Rothberg et al. (2011)]