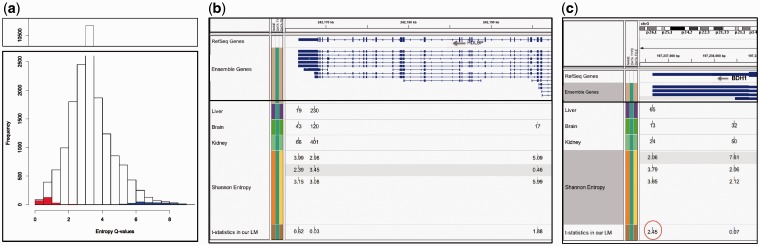
Fig. 3.
(a) Histogram of Shannon entropy Q-values for all PAS clusters (range from 0 to 9). Red bars represent entropy values for individual-significant PAS detected by our model, blue bars represent overlap-significant PAS. Individual sites cluster at 0–2 entropy, and overlap sites cluster at the upper end of the range. (b) Example showing that Shannon entropy does not take the abundance of the evidence into account for calling sites significant. The usage (count) of each PAS cluster is marked in each tissue, followed by the entropy values then the test statistics resulted from our linear model. Tissue specificity is determined by low entropy values but high test statistics above a certain threshold. The proximal site in brain (17 tags;  of total) is classified as specific (entropy = 0.46). However, this relatively low tag number compared with the overall expression of the gene and the total library depth is not enough to call this site brain-specific. Entropy values for this proximal site in liver and kidney represent pseudo-counts (not shown in figure). (c) Example of a specific PAS cluster detected by our linear model and not by Shannon entropy (BDH1 gene on negative strand). The distal site (65 tags) is the only PAS site for this gene used in liver. Given the relatively low expression level of the gene and the liver-low library depth compared with brain and kidney, this site is classified as significant (test statistic of our model is marked by red circle). Shannon entropy values do not reflect this relative usage
of total) is classified as specific (entropy = 0.46). However, this relatively low tag number compared with the overall expression of the gene and the total library depth is not enough to call this site brain-specific. Entropy values for this proximal site in liver and kidney represent pseudo-counts (not shown in figure). (c) Example of a specific PAS cluster detected by our linear model and not by Shannon entropy (BDH1 gene on negative strand). The distal site (65 tags) is the only PAS site for this gene used in liver. Given the relatively low expression level of the gene and the liver-low library depth compared with brain and kidney, this site is classified as significant (test statistic of our model is marked by red circle). Shannon entropy values do not reflect this relative usage