Figure 1.
Antiviral Immune Responses Analogous to Bacterial and Fungal Immune Responses.
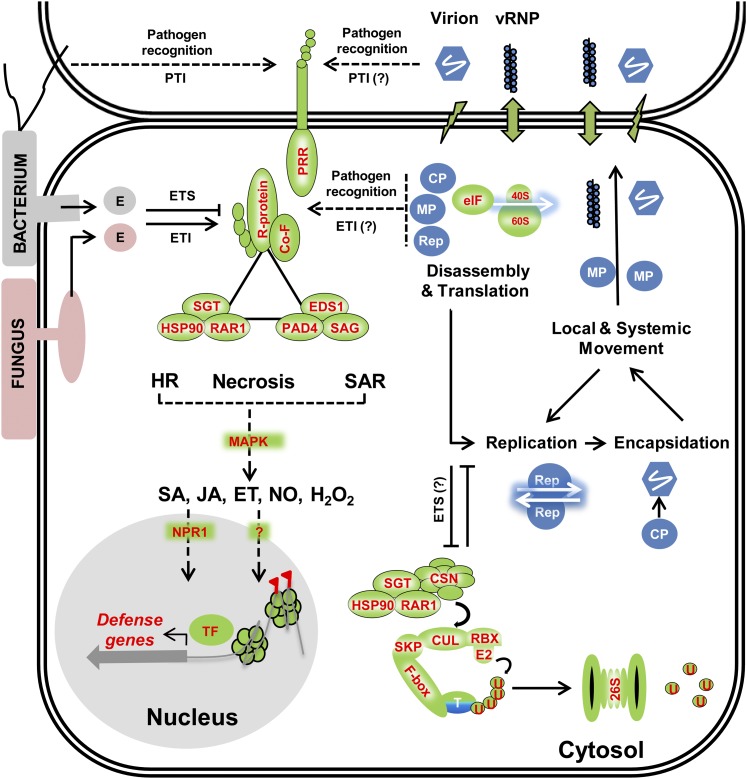
Viruses typically enter plant cells through cellular damage (lightning bolt) and move from cell to cell primarily via plasmodesmata (up-down arrow) as viral ribonucleoprotein complexes (vRNP) and/or virions. Virus-encoded proteins such as replicase (Rep), capsid protein (CP), and movement protein (MP) are translated within the host cell cytosol and cooperatively function in translation, replication, encapsidation, and movement of the virus. Similar to events that occur during bacterial and fungal-triggered immune responses, virus-associated factors, such as virion components or virus-encoded proteins, could be perceived by putative cell surface PRRs or cytosolic NB-LRR receptors (e.g., R proteins) to trigger analogous ETI or susceptible (ETS) responses, culminating in HR, SAR, and/or necrosis phenotypes. Bacterial and fungal secreted effector proteins involved in ETI signaling are indicated by “E.” In a manner similar to bacterial and fungal ETI responses, virus-triggered ETI responses also involve functional SGT1/RAR1/HSP90 (Liu et al., 2004) and EDS1/PAD4/SAG101 (Zhu et al., 2011) protein complexes. Combinatorial interactions between viral proteins, R proteins, R cofactors (Co-F), SGT1/RAR1/HSP90, and EDS1/PAD4/SAG101 complexes mediate distinct downstream changes in SA, JA, ethylene (ET), NO, and hydrogen peroxide levels or signaling via MAP kinase signaling cascades. NPR1, a nucleo-cytoplasmic protein critical for nonviral SA defense responses, mediates transcriptional changes in defense gene expression via interactions with specific transcription factors (TFs) (Dong, 2004). The majority of virus-triggered SA responses, however, appear to be NPR1 independent (Whitham et al., 2003). In addition to TF-mediated transcriptional changes, viruses also trigger chromatin modifications including DNA methylation changes (red flag) and increased homologous recombination rates (Kovalchuk et al., 2003; Boyko et al., 2007; Kathiria et al., 2010), epigenetic changes that could be stable inherited by progeny. The SGT1/RAR1/HSP90 complex also interacts with CSN components to mediate degradation of either viral or host proteins (T) via 26S-proteosome complex (Liu et al., 2002b; Shirasu, 2009). The 26S proteasome also functions in the nucleus, aspects that are discussed in detail elsewhere (Padmanabhan and Dinesh-Kumar, 2010). Ubiquitin protein is abbreviated as “U.” Unknown or putative paradigms are indicated as “?.”