Figure 1.
ETPamir Screens for Single Gene Silencing in Arabidopsis.
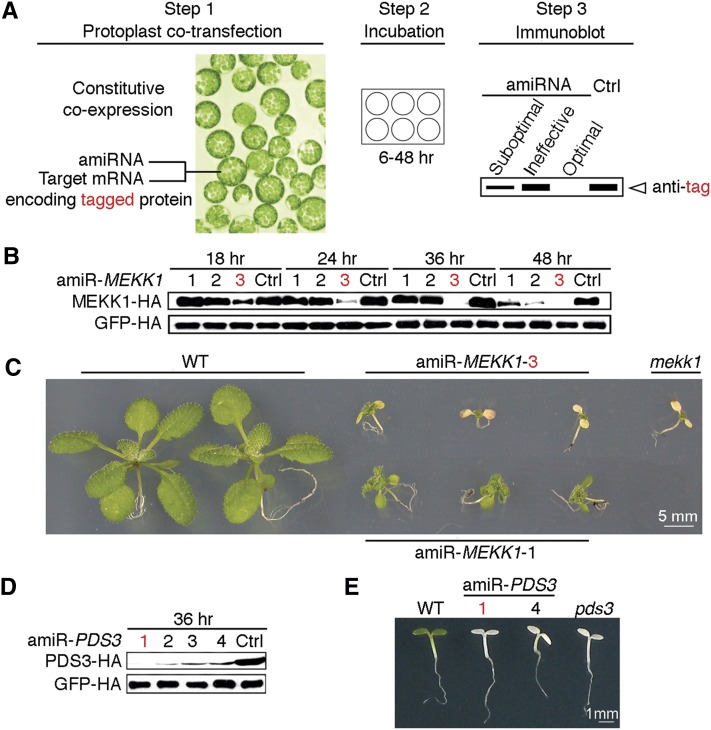
(A) Scheme of the ETPamir screen. The use of an epitope tag and immunoblot analysis by an antitag antibody for target protein quantification are highlighted in red. The time of protoplast incubation depends on the target protein stability. Empirically, 36 h after cotransfection is an optimal time point to discriminate optimal, moderate, and ineffective amiRNAs for most target genes. Unstable target proteins require shorter incubation time (e.g., 6 to 12 h). Ctrl, control.
(B) Time-course immunoblot of MEKK1-HA protein to define optimal amiR-MEKK1. The optimal amiRNA, amiR-MEKK1-3, is highlighted in red.
(C) Transgenic Arabidopsis plants overexpressing amiR-MEKK1-3 resemble the mekk1 null mutant (SALK_052557). WT, the wild type.
(D) Immunoblot of PDS3-HA protein to define optimal amiR-PDS3. The most efficient amiRNA, amiR-PDS3-1, is highlighted in red.
(E) Transgenic Arabidopsis plants overexpressing amiR-PDS3-1 or amiR-PDS3-4 resemble the pds3 null mutant (SALK_060989).
The numerical order of each amiRNA was based on the high-to-low WMD ranking. In (B) and (D), three independent repeats with GFP-HA as an untargeted internal control produced similar results.