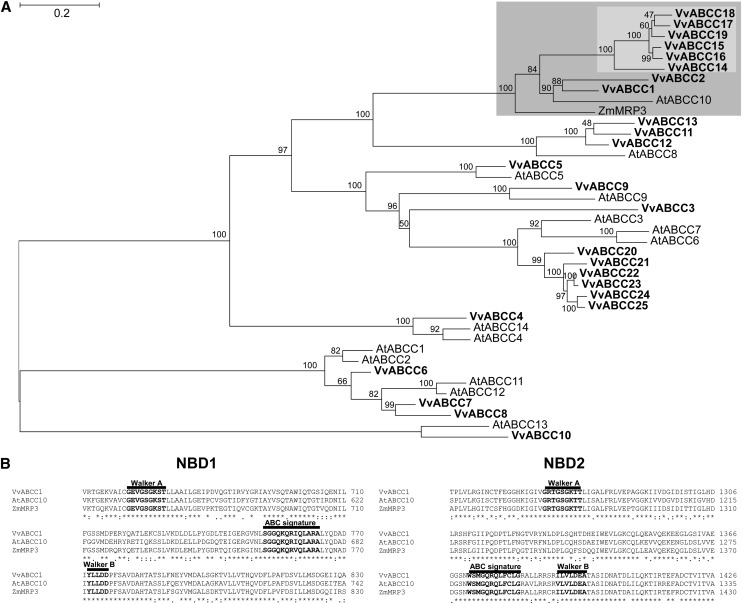
Figure 1.
Phylogeny of Grapevine ABCC Proteins.
(A) Maximum likelihood phylogeny of grapevine (Vv), Arabidopsis (At) ABCC-type proteins, and Zm-MRP3/Zm-ABCC3. The clade comprising Vv-ABCC1, Vv-ABCC2, and Zm-MRP3 is shaded in dark gray, and within this clade, the subclade comprising the six other Vv-ABCCs is shaded in light gray. Branch support values indicate nonparametric bootstrap values (in percentages of 1000 replicates). The tree was estimated with PhyML 3.0 from a trimmed protein sequence alignment.
(B) Predicted ABC-type motives in Vv-ABCC1, At-ABCC10, and Zm-MRP3. The Walker A and B motifs as well the ABC signature are shown in bold letters. The three-character code is used to highlight conserved amino acids: *, full identity; :, one of the following strong groups is fully conserved (STA, NEQK, NHQK, NDEQ, QHRK, MILV, MILF, HY, and FYW); ., one of the following weaker groups is fully conserved (CSA, ATV, SAG, STNK, STPA, SGND, SNDEQK, NDEQHK, NEQHRK, FVLIM, and HFY). NBD, nucleotide binding domain. Bar = 0.2 amino acid substitutions per site.