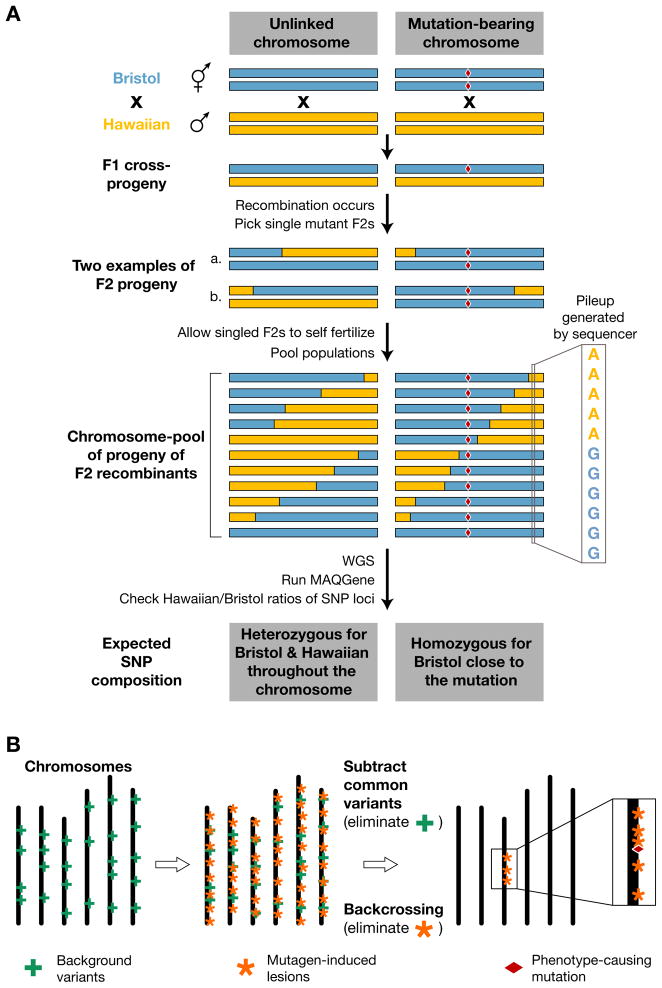
Figure 1. Mutant mapping by whole genome sequencing.
(A) WGS-SNP mapping. A homozygous mutant strain (mutation denoted by a red diamond) in a Bristol background is crossed with a SNP-containing Hawaiian strain. Mutant worms segregating from this cross are reselected in the F2 generation. These mutant lines carry chimeric chromosomes (containing Bristol and Hawaiian SNPs) generated by meiotic recombination. When DNA from multiple F2 recombinant lines is pooled and sequenced, then the relative number of Bristol vs. Hawaiian SNP at a given location in the genome will be a reflection of the relative distribution of recombinants in the pool. Therefore, only Bristol SNPs will be found close to the mutation of interest (adapted from Ref. 28).
(B) Mapping based on mutagen-induced DNA variation density across the genome. Mutagenized strains contain not only mutagen-induced damage (orange asterisks) but also carry background variants (green crosses) present in the mother strain before mutagenesis. By backcrossing a candidate mutant line to the strain used for the original mutagenesis, it is possible to eliminate most mutagen-induced lesions except for those which are closely linked to the phenotype-causing mutation (red diamond). In addition, background variation present in the unmutagenized strain can by identified by comparing independent backcrossed mutant lines. After filtering, the genomic region linked to the phenotype-causing mutation will be highlighted by a high-density cluster of mutagen-induced lesions (adapted from Ref. 29).