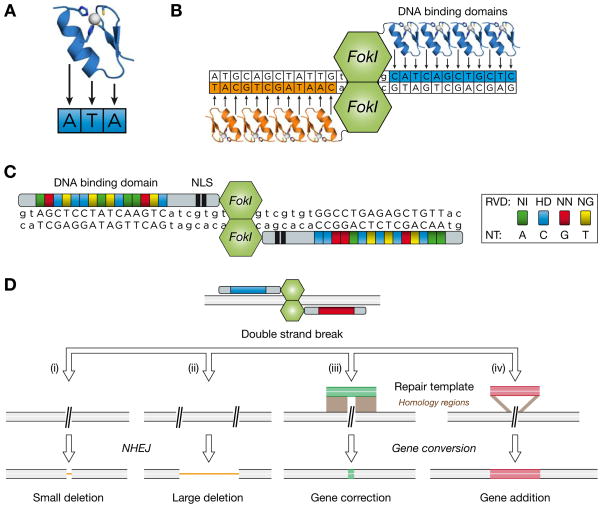
Figure 4. Genome editing with engineered nucleases.
(A) Structure of a Zinc Finger (ZF) DNA-binding module. Each individual ZF domain is optimized to recognize a three base pair sequence. Two cysteines and two histidines coordinate a zinc atom in each module.
(B) ZF nucleases (ZFN) are composed of at least four ZF domain repeats, linked to the FokI endonuclease domain. Specificity is increased by using obligatory dimers of two ZFN recognizing two sites which flank the targeted double-strand break site.
(C) Structure of TALE nucleases (TALEN). A central array of tandem repeats forms the specific DNA binding domain. Two nucleotides (repeat-variable di-residues, RVD) in each repeat determine the sequence preference of each module. Examples of four RVDs that recognize all four nucleotides (NT) are indicated. Two TALEN monomers bind to their target sites and allow the dimerization of the FokI endonuclease domain. TALENs also contain a nuclear localization signal (NLS).
(E) ZFNs and TALENs allow different types of genome editing strategies. A double strand-break (DSB) can be resolved by different pathways. (i) A single DSB can be repaired by non-homologous end-joining (NHEJ) and result in a small deletion of a few base pairs. Alternatively, (ii) two simultaneous DSB can generate a larger deletion allele. When a repair template with homology regions is provided, the DSB is resolved by gene conversion to (iii) insert a small sequence (as small as a single nucleotide, i.e. “gene correction”) or (iv) a larger transgene (“gene addition”).