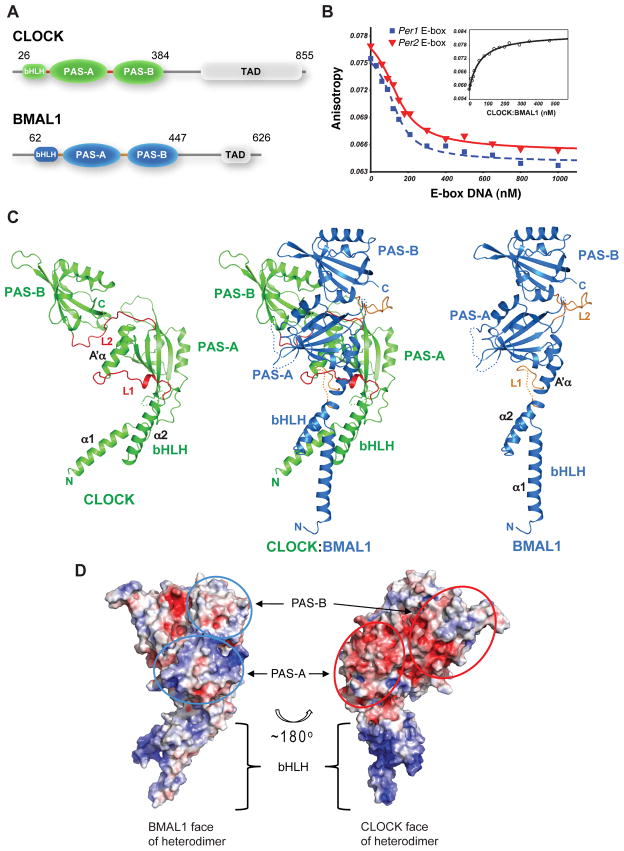
Fig. 1.
Overall structure of mouse CLOCK:BMAL1. (A) Domain organization of CLOCK and BMAL1. Crystals were obtained from the truncated proteins (indicated by the amino acid residue number) encompassing the bHLH-PAS-AB domains. (B) DNA-binding affinity of the truncated CLOCK:BMAL1 complex measured by fluorescence anisotropy. Dissociation constant (Kd) of the fluorescein labeled mPer2 E2-box DNA was 59 ± 7.3 nM by direct binding to CLOCK:BMAL1 (inset). Using unlabeled DNA probes as competitor, the Kd’s of unlabeled 18-mer mPer1 E1-box DNA (blue) and mPer2 E2-box DNA (red) (40) were 9.0 ± 2.3 nM and 13 ± 2.0 nM, respectively. (See Materials and Method for details). (C) Ribbon diagram of CLOCK:BMAL1 heterodimer (center). The CLOCK subunit is colored green, BMAL1 blue. Each individual domain is labeled. The CLOCK (left) and BMAL1 (right) subunits are also shown separately to illustrate their different spatial domain arrangements. The linker regions between domains in the two subunits (L1 and L2) are highlighted in red or orange color. Flexible loops lacking density are indicated by dotted lines. (D) Electrostatic potentials of CLOCK:BMAL1 heterodimer showing that the surfaces composed of CLOCK PAS domains (red ovals, right) have mostly negative potentials while the surfaces of BMAL1 PAS domains (blue ovals, left) are mostly positive or neutral. The colors are ramped from negative potential −5 kT/q (red) to positive 5 kT/q (blue).