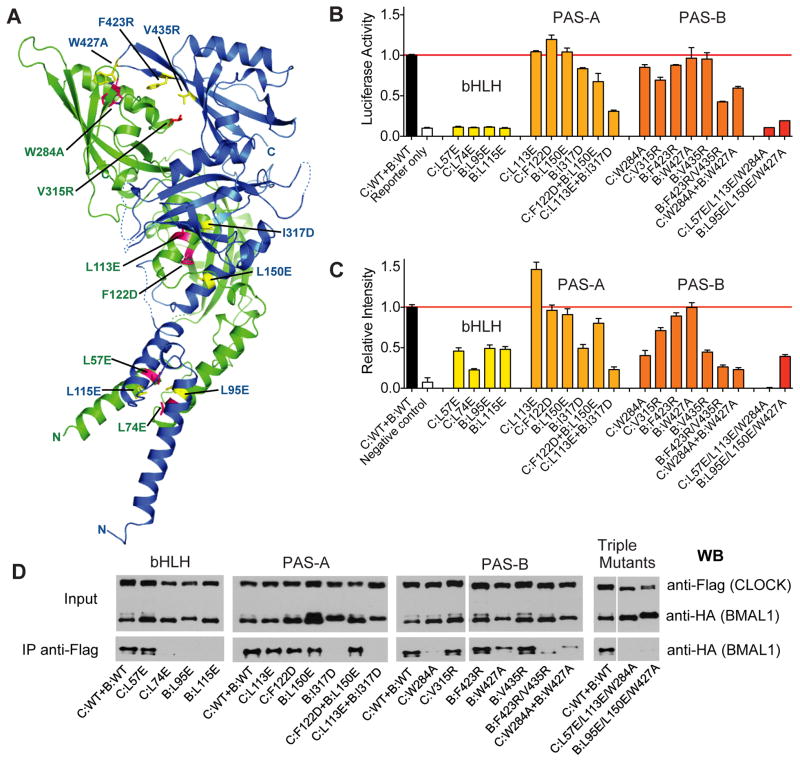
Fig. 4.
Functional analysis of CLOCK:BMAL1 mutants. (A) Locations of domain interface mutants in CLOCK (green) and BMAL1 (blue). (B) Per2 promoter:Luciferase reporter assays to evaluate the effects of structure-based mutations on transactivation by full-length CLOCK:BMAL1. Data are average of 2 independent experiments performed in duplicate. (C) Bimolecular fluorescence complementation (BiFC) experiments on the same set of mutants in CLOCK:BMAL1 truncated constructs. The fluorescent intensities of WT and mutant CLOCK:BMAL1 bHLH-PAS-AB constructs (for details, see Supplementary Materials and Methods) were quantified using data from 3 independent experiments. (E) Coimmunoprecipitation experiments assessing the association of CLOCK and BMAL1 in full-length WT and mutant proteins. Anti-FLAG affinity gel was used to precipitate FLAG tagged CLOCK along with the tightly associated BMAL1, which is HA tagged. The Western blots using an anti-HA antibody were then performed to detect the association of WT and mutant BMAL1 with CLOCK constructs. The Co-IP data are representative of at least 3 independent replicates, with the exception of C:W284A which had stronger co-IP interaction in other experiments, but on average was weaker than WT CLOCK.