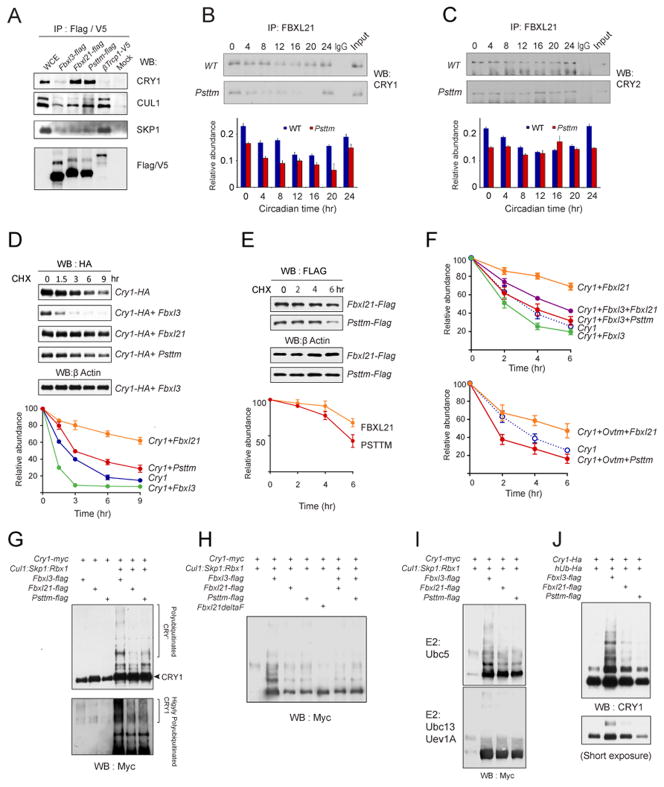
Figure 4. FBXL21 forms an SCF E3 ligase complex that slowly ubiquitinates CRY1 and antagonizes the activity of SCFFbxl3.
A. Interaction of FBXL proteins with Cullin1 and Skp1. NIH3T3 cells were transfected with Flag-Fbxl3, Flag-Fbxl21, Flag-Psttm and V5-βTrcp1 expression constructs. Co-immunoprecipitated proteins were analyzed by western blotting with anti-CRY1, anti-CUL1, and anti-SKP1 antibodies. Representative blots from 3 independent experiments are shown.
B and C. FBXL21 interacts with CRY1 (B) and CRY2 (C) in native extracts in a circadian manner. Liver extracts from CT0 to CT24 were immunoprecipitated (IP) with an FBXL21 antibody, and Western blotting was performed with CRY1 and CRY2 antibodies. Lower: quantification of co-immunoprecipitated CRY1 and CRY2 amounts. Data are taken from 2 independent experiments. Error bars show ± range (n = 2). 2-way ANOVA shows statistically significant differences between WT and Psttm for the amount of CRY1 co-precipitated with FBXL21 throughout the circadian cycle (p < 0.0001).
D. Differential effects of FBXL21 and PSTTM on CRY1 stability. 293A cells were co-transfected with indicated constructs. 32 hrs after transfection, cells were treated with 20 μg/ml cycloheximide and incubated for the indicated time before harvest. Western blotting was performed to monitor CRY1 levels using an anti-HA antibody. Lower: Quantification of the effects of FBXL3, FBXL21, PSTTM on CRY1 stability. Data are taken from 3 independent experiments. Error bars show ± SEM (n = 3). Half life was determined by using nonlinear, one phase exponential decay analysis (the half-life parameter, K, is significantly different in all four conditions: p<0.0001)
E. The Psttm mutation destabilizes FBXL21. 293A cells were transfected with Fbxl21-Flag or Psttm-Flag constructs. Cycloheximide treatment was performed as in D. Data are taken from 3 independent experiments. Error bars show ± SEM (n = 3). The half-life parameter, K, is significantly different: p=0.0436.
F. Competition between FBXL3 and FBXL21/PSTTM modulates CRY1 degradation. Cycloheximide treatment and CRY1 western blotting were performed as in D. Data for quantification represent 3 independent experiments (Figure S4F). The half-life parameter, K, is significantly different: p<0.0001. Data are taken from 3 independent experiments. Error bars show ± SEM (n = 3).
Lower: Competition between Ovtm and Fbxl21/Psttm in CRY1 degradation. Cycloheximide treatment and CRY1 western blotting were performed as in D. Data for quantification represent 3 independent experiments (representative western blots are shown in Figure S4G). The half-life parameter, K, is significantly different: p<0.0001. Error bars show ± SEM (n = 3).
G. In vitro ubiquitination assay. Sf9 cells were infected with the indicated baculovirus constructs. Samples were analyzed by western blotting with an anti-Myc antibody. Upper shows a short exposure image, and the bracket to the right marks polyubiquitinated CRY1. Lower shows a long exposure image, and the bracket to the right marks highly polyubiquitinated CRY1. Results are representative of more than 3 replicates.
H. In vitro ubiquitination was performed as indicated in G. Sf9 cells were infected with the indicated baculovirus. Co-infection of Fbxl baculovirus (Fbxl3+Fbxl21, Fbxl3+Psttm) attenuated the E3 ligase activity of FBXL3. Results are representative of 3 replicates.
I. FBXL3/FBXL21 mediated CRY1 ubiquitination requires Ubc5 as an E2 ligase. Top: Ubc5-mediated robust ubiquitination by FBXL3-SCF complexes and multi ubiquitination by FBXL21/PSTTM SCF complex. Lower: lack of CRY1 ubiquitination in the presence of Ubc13/Uev1A as the E2 ligase. Results are representative of 3 replicates.
J. 293A cells were co-transfected with Cry1-HA, ubiquitin (hUb-HA) and the indicated F-box constructs. Cells were treated with MG132 (10μg/ml) 6 hr before harvest. Whole-cell lysates were analyzed by western blotting with an anti-CRY1 antibody. Results are representative of 3 replicates. See also Figure S4.