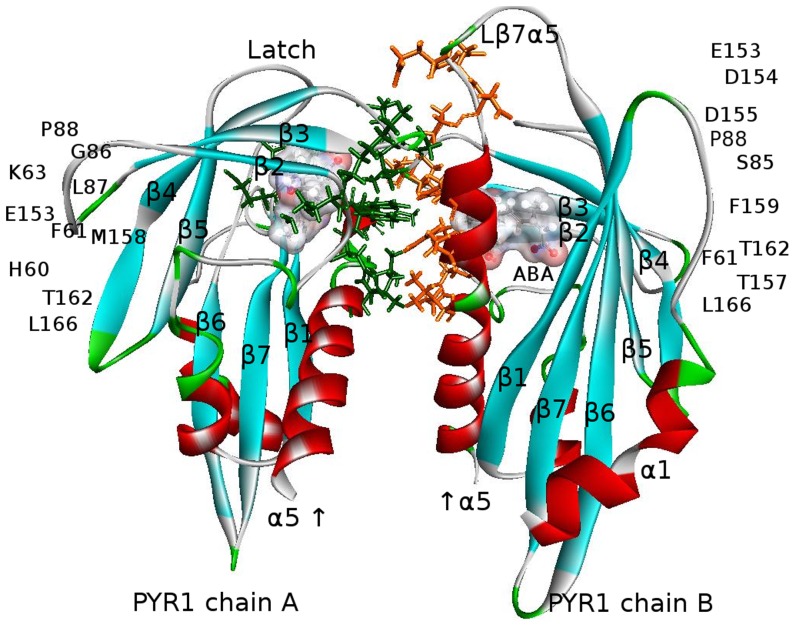
Figure 10. PYR1-dimer, 2ABA -bound (modified PDB ID 3NJO after ligand replacement, mutation S88P, minimizations and equilibrations in water) with residues on the binding surface indicated by orange sticks for chain A and green sticks for chain B.
The direct and water-mediated interactions, detected by AccelrysVS employing the same criteria as for the PYR1-ABA-HAB1 complex in Figure 6, comprise H60–L166, H60–T162, F61–F159, F61–L166, F61–F61, F61–T162, I62–M158, K63–D155, K63–E153, I84–F159, S85–D155, S85–D154, S85–F159, S85–T156, S85–E153, G86–P88, G86–L87, G86–F159, L87–P88, L87–L87, L87–F159, P88–P88, L166–L166 and all reciprocal [10]. Ligand molecules in the binding pockets are depicted by surfaces colored according to the charge distribution as in Figure 6.