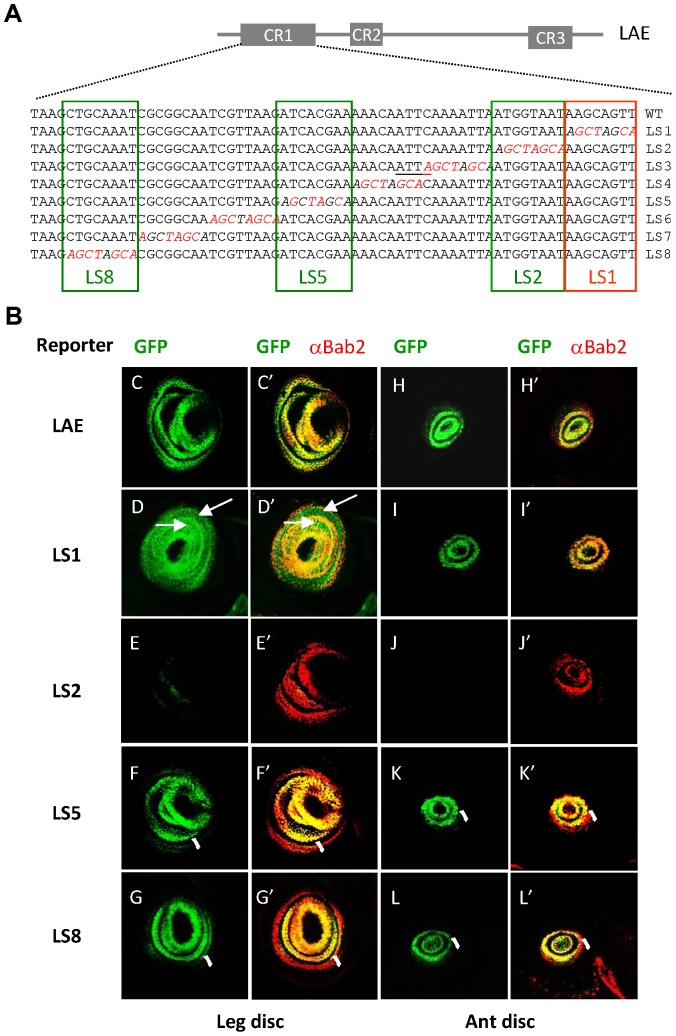
Figure 4. The key CR1 sequence includes separate activating and repressive regulatory information.
(A) Linker scanning (LS) mutagenesis of the 68-bp CR1 conserved region. The sequences of the wild-type and of each of the eight mutated (LS1–8) CR1 sub-regions (tested within the entire LAE) are shown, with the names of the examined LS constructs indicated on the right side. The inserted linker sequence (AGCTAGCA) is italicized with nucleotide substitutions depicted in red. Note that an additional consensus HD-binding site (underlined) has been created in the LS3 construct. The most striking positive and negative regulatory elements are framed in green and red, respectively. (B) Critical DNA elements within CR1. GFP expression (green) alone and in combination with Bab2 immunostaining (red) of leg (C–G and C′–G′, respectively) and antennal (ant) (H–L and H′–L′, respectively) imaginal discs from late third-instar larvae, expressing either the LAE-GFP construct or any of the four LS-mutated derivatives displaying the strongest abnormal expression patterns, are shown (for the remaining four constructs, see Figure S4). LS2-driven GFP reporter expression was drastically reduced, in both leg (E–E′) and antennal (J–J′) imaginal discs. Conversely, the LS1-GFP construct displayed inter-ring de-repression in leg tissues (D–D′), as determined by the green (arrows) instead of yellow colour normally seen in the merged images. Finally, the LS8 and to a lesser extent LS5 constructs exhibited strongly decreased GFP expression in the proximal-most bab2-expressing tarsal (F–G) and antennal (K–L) as determined by the red staining (brackets) instead of the yellow colour normally seen on the merge.