FIGURE 6.
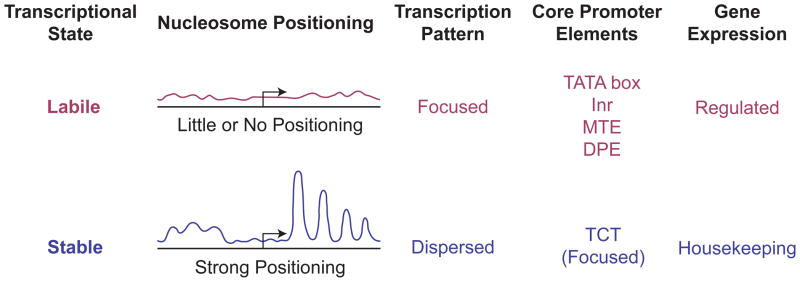
Chromatin structure and transcriptional state. Promoters can be viewed from the perspective of the transcriptional state of the gene. In the case of housekeeping genes, the transcriptional state is stable and the flow of factors through the promoter region can be viewed as that of a steady state system. This stable transcriptional state correlates with an ordered chromatin structure in the promoter region, wherein a positioned array of nucleosomes is typically observed. Regulated genes are in a labile transcriptional state in which they are poised for rapid activation or repression. This labile state correlates with little or no apparent order in the chromatin structure in the promoter region. Focused promoters, particularly those with TATA, Inr, MTE, or DPE motifs, are generally associated with regulated genes and a lack of positioned nucleosomes in the promoter region. Dispersed promoters, on the other hand, are commonly found in housekeeping genes with positioned nucleosomes in the promoter region. One notable exception to this general trend is the TCT motif-based transcription system, in which focused promoters mediate the transcription of ribosomal protein genes, which are housekeeping genes. Unlike the TATA-, Inr-, MTE-, and DPE-containing promoters, TCT-dependent promoters exhibit strong nucleosome positioning.31 These findings suggest that the key feature that dictates the chromatin structure in the promoter region is the stability or lability of the transcriptional state.
