Figure 4.
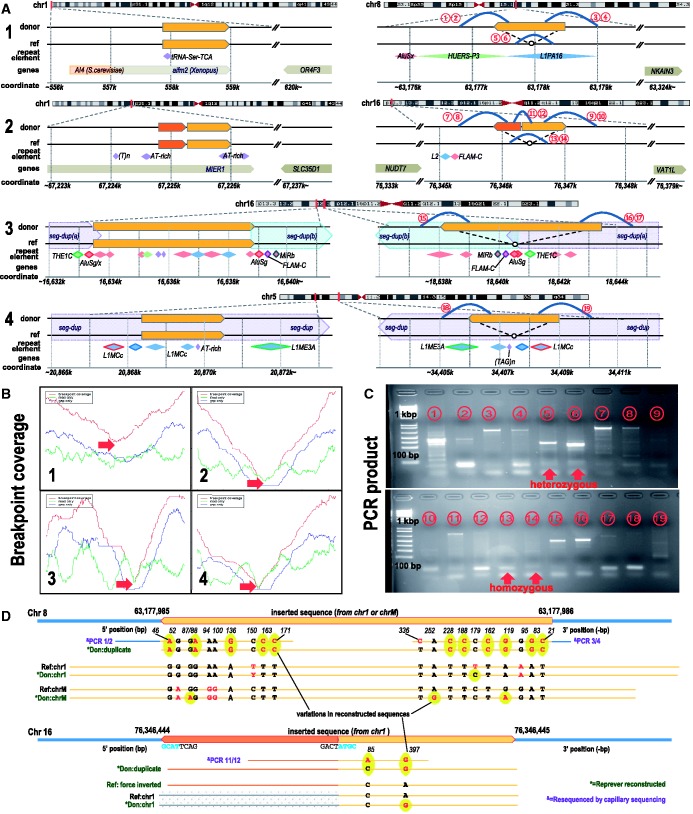
(A) Four cases are illustrated as examples. (1) A gain call is made at chr1:553585-560412 by CNVer (and chr1:554301–560300 by Yoon et al.). Reprever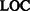 found that a ∼1.3-kb sub-region is duplicated into chr8:63177985. (2) Similarly, a gain call at chr1:67224800–67226000 by Yoon is explained by Reprever
found that a ∼1.3-kb sub-region is duplicated into chr8:63177985. (2) Similarly, a gain call at chr1:67224800–67226000 by Yoon is explained by Reprever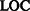 showing a duplication at chr16:76346444. Note that the inserted sequence has a partial inversion of 400-bp 5′ segment (orange block). (3 and 4) Two gain calls were made CNVer (chr16:16633477–16638823 and chr5:20868352–20870585). Insertion locations found by Reprever
showing a duplication at chr16:76346444. Note that the inserted sequence has a partial inversion of 400-bp 5′ segment (orange block). (3 and 4) Two gain calls were made CNVer (chr16:16633477–16638823 and chr5:20868352–20870585). Insertion locations found by Reprever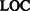 are located in bigger segmental duplication regions (dotted blue/red blocks). The arrangement of repeat elements (colored diamonds) show that the copy number increase in the donor genome actually resulted from deletions in the reference genome. (B) Concordant read counts around the putative breakpoints (red arrows). Coverage is calculated using whole-clone (red), end reads (green) and the gap between end reads (blue) of mate pairs. The relative read count reduction predicts the allele zygosity (1/3 heterozygous and 2/4 homozygous). (C) PCR amplification of duplicated regions. Primers are designed to capture duplication boundaries (encircled red numbers in A). (D) (upper) The inserted sequence of the first case (at chr8:63177988) is identified by Sanger sequencing (annotated with PCR 1/2 and PCR 3/4). Totally 12 SNVs are found. Three consensus sequences reconstructed by Reprever
are located in bigger segmental duplication regions (dotted blue/red blocks). The arrangement of repeat elements (colored diamonds) show that the copy number increase in the donor genome actually resulted from deletions in the reference genome. (B) Concordant read counts around the putative breakpoints (red arrows). Coverage is calculated using whole-clone (red), end reads (green) and the gap between end reads (blue) of mate pairs. The relative read count reduction predicts the allele zygosity (1/3 heterozygous and 2/4 homozygous). (C) PCR amplification of duplicated regions. Primers are designed to capture duplication boundaries (encircled red numbers in A). (D) (upper) The inserted sequence of the first case (at chr8:63177988) is identified by Sanger sequencing (annotated with PCR 1/2 and PCR 3/4). Totally 12 SNVs are found. Three consensus sequences reconstructed by Reprever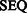 (green) recovered 11 of 12 variations (92%) for inserted sequence and found four additional variations in the homologs (highlighted in yellow). (lower) The partial inversion of the inserted segment in the second case (at chr16:76346444) is confirmed by Sanger sequencing with two SNVs. Reprever
(green) recovered 11 of 12 variations (92%) for inserted sequence and found four additional variations in the homologs (highlighted in yellow). (lower) The partial inversion of the inserted segment in the second case (at chr16:76346444) is confirmed by Sanger sequencing with two SNVs. Reprever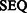 recovered one of them.
recovered one of them.