Figure 1.
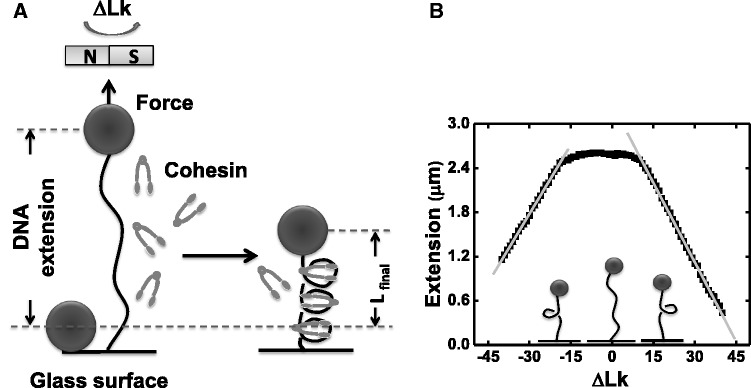
Experimental design, calibration of DNA extension versus ΔLk (A) An 9.8 kb DNA linearized pFos-1 is attached at one end to glass cover slip, the other to a 2.8 µm magnetic bead (filled circles). Upon protein flow through, the DNA length change owing to cohesin/DNA interaction can be detected. (B) Calibration data for DNA extension as a function of linking number change (ΔLk) under 0.45 pN force, in buffer with no protein present. At zero supercoiling, the tether extends to ∼2.7 µm. For <12 turns, there is little or no change in DNA extension. Beyond 12 turns, the DNA molecule buckles and plectonemic regions are generated, resulting in a simple linear reduction of DNA extension with ΔLk (for ΔLk > 11, slope is −62.3 ± 0.4 nm). For ΔLk < −14 turn, molecule buckles and the slope of the linear plectonemic regions is nearly the same as that of positive supercoiling. (for ΔLk < −14, slope is 71.2 ± 0.5 nm).
