Abstract
Inhibitors of histone deacetylases (HDAC) are an important emerging class of drugs for the treatment of cancers. HDAC inhibitors are currently under evaluation in clinical trials as single agents and as sensitizers in combinations with chemotherapies and radiation therapy. Although these drugs have important effects on cancer cell growth and functions, the mechanisms underlying HDAC inhibitor activities remain to be fully defined. By using rational drug design, compound 2, a fluorescent class II HDAC targeting inhibitor, was synthesized and observed to accumulate in the cytoplasmic compartments of treated cells, but not in the nuclei. Furthermore, immunostaining of inhibitor exposed cells for HDAC4 showed accumulation of this enzyme in the cytoplasmic compartment with concomitant increased acetylation of tubulin and nuclear histones. These observations support a mechanism by which nuclear histone acetylation is increased as a result of HDAC4 trapping and sequestration in the cytoplasm after binding to compound 2. The HDAC inhibitor offers potential as a novel theranostic agent, combining diagnostic and therapeutic properties in the same molecule.
Introduction
The current understanding of the roles of acetylation in the epigenetic regulation of chromatin structure and gene expression rests on the balance of activities of histone acetyltransferases and histone deacetylases (HDAC). Increased acetylation of histones leads to changes in chromatin structure and accessibility of key cellular proteins to specific target sites (1, 2). Histone acetyltransferases’ acetylate lysine groups at the amino terminal tails of nuclear histones neutralize positive charges on the histones, yielding a more open, transcriptionally active chromatin structure (3). In contrast, the HDACs deacetylate and suppress transcription. In this model, inhibitors of HDACs bias the balance toward a more acetylated state. Such a shift in the relative activities of these enzymes may change gene expression necessary for DNA repair, replication, cell cycle checkpoint activation, and tumor suppression (4, 5).
Human histone deacetylases can be divided into 4 classes based on structure, sequence homology, and domain organization. Class I consists of HDACs 1, 2, 3, and 8. Class I HDACs are nuclear and play roles in cell proliferation and apoptosis. Class II includes HDACs 4, 5, 6, 7, 9, and 10 (6). These enzymes are characterized by a large NH2-terminal domain or a second catalytic site and their expression is more restricted, suggesting roles in cellular differentiation and development (2). Class III enzymes include the sirtuins and are NAD-dependent deacetylases (7). Class III enzymes are not inhibited by TSA or other hydroxamates. HDAC11 is designated as class IV based on phylogenetic analysis (8).
HDACs are found in the nuclear and cytoplasmic compartments. Although they are involved in critical cellular functions, such as cell cycle regulation and apoptosis, a key function of HDACs is transcriptional regulation. HDACs function as components of large multiprotein complexes that bind to promoters and repress transcription. Class I HDACs are predominantly sublocalized in the nucleus, whereas class II HDACs shuttle between the nucleus and the cytoplasm; however, both classes of HDACs have conserved deacetylase core domains of approximately 400 amino acids and zinc-binding sites. It is the core domain that presents the principal target for design of inhibitory small molecules. Key reported acetylation mechanisms relevant to cancer treatment have involved histones and tubulin, as well as a variety of other nonhistone proteins (9–12). We have reported radiation sensitizing properties of novel hydroxamic acid and mercaptoacetamide inhibitors in a search for HDAC isomer specificity (13, 14). By designing and synthesizing new class II targeting HDAC inhibitors offering intrinsic fluorescent properties, we provide further insight into action mechanisms and subcellular sites of inhibitor activities.
Materials and Methods
Materials
HeLa nuclear extracts and fluorimetric histone deacetylase assay kits were obtained from BIOMOL. Antibodies for acetylated a-tubulin and HDAC4 were purchased from Sigma, acetylated histone H4 from Upstate Biotechnology, and glyceraldehyde-3-phosphate dehydrogenase from Trevigen.
As a quality control and to confirm the identity of the cultured cell lines used in these experiments, cell lines were submitted to genetic fingerprinting. The commercially available Cell IDTM System (Promega) was used to screen cell lines obtained from the Georgetown Tissue Culture Shared Resource (LNCaP). Cells that were obtained from the American Type Culture Collection (A549, MCF7, C42, and PC-3) were genetically fingerprinted by using the PowerPlex 1.2 System by the American Type Culture Collection and had been passaged for less than 6 months before use in these experiments. All cell lines were screened for the presence of mycoplasmids before use by the Georgetown Tissue Culture Shared Resource by using the Gen-Probe Mycoplasma Tissue Culture NI Rapid Detection System.
Synthesis
Schemes, methods of syntheses, and spectroscopic data for the new compounds are presented in the Supplementary Material. Purity and identity were established for all compounds by using mass spectrometry, NMR spectrometry, and high-performance liquid chromatography analyses.
HDAC assays
The values of IC50 concentrations of HDAC inhibitors were determined by carrying out a fluorimetric histone deacetylase assay following the manufacturer’s instructions. For the pan-HDAC assay, HeLa nuclear extracts were used as a source of histone deacetylase, and the histone deacetylase isomer inhibition assays were done by using purified recombinant histone deacetylase proteins of the various isomers (BPS Bioscience Inc.). Reactions were prepared in 0.1 mol/L KCl, 20 mmol/L HEPES/ NaOH at pH 7.9, 20% glycerol, 0.2 mmol/L DTA, 0.5 mmol/L DTT, and 0.5 mmol/L phenylmethylsulfonyl-fluoride. The HDAC assay was done by using Fluor de Lys substrate and concentrations (nmol/L to µmol/L) of compounds at 37°C in HDAC assay buffer, containing 25 mmol/L Tris-Cl, pH 8.0, 137 mmol/L NaCl, 2.7 mmol/L KCl, and 1 mmol/L MgCl2. Fluorescence was measured with an excitation at a wavelength of 360 nm and emitted light of 460 nm (TECAN ULTRA 384) was detected. Negative (no enzyme, no inhibitor, a drug with no HDAC inhibition activity) and positive controls (a HeLa nuclear extract with no HDAC inhibitor and known HDAC inhibitor SAHA) were included in the assay reactions. The reaction was done in triplicate for each sample. Each point represents the mean ± SD of replicates.
Cell growth inhibition assay
Cell lines PC-3 (prostate cancer), C42 (androgen-independent prostate cancer), and LNCaP (androgen-dependent prostate cancer) were cultured in RPMI-1640 medium supplemented with 100 mg/mL of streptomycin, 100 mg/mL of penicillin, 2 mmol/L L-glutamine, 1 mg/mL of hydrocodisone, and 20% FBS. These cultures were maintained in a 95% air/5% CO2 incubator at 37°C After 24 hours seeding, a solution of the compound or dimethyl sulfoxide (DMSO) vehicle control were added to cells and incubated for 48 hours at 37°C. Growth inhibition was determined by using the CellTiter 96 AQueous One Solution Cell Proliferation Assay according to manufacturer’s instructions (Promega, Inc.). The absorbance was measured at 490 nm on a microplate reader. The 50% growth inhibition (GI50) was calculated as the compound concentration required reducing cell number by 50% compared with control.
Cell cycle distribution
LNCaP and PC-3 cells were incubated in the presence or absence of compound before harvesting at the indicated time points, washed twice with cold PBS, and 1 × 106 to 2 × 106 cells were resuspended in 0.5 mL of ice-cold PBS for analysis. Single-cell suspensions were verified microscopically. The cell suspension was gradually mixed with ice-cold absolute ethanol to the final volume of 2.0 mL and cooled on ice at least for 20 minutes before staining with propidium iodide. The cell cycle distributions were measured in a fluorescence-activated cell sorter from Becton Dickinson. DNA content levels were quantified by using ModFit LT 3.0 software (Verity Software House, Inc.).
Fluorescent measurement
Cells were plated onto glass slides at a density of 500,000 per slide and incubated overnight. Cells were replenished with a medium containing 20 µmol/L of compound 2 and incubated for 1 hour. Slides were washed with PBS, cells were fixed with a 4% solution of formaldehyde for 10 minutes, washed, and mounted. Nuclear staining was achieved by treating cells with propidium iodide (1:1,000) for 4 minutes after fixation. The fluorescent compound was imaged by using a multiphoton laser with an excitation wavelength of 700 nm and an emission wavelength of 510 nm. Propidium iodide was detected with an excitation wavelength of 535 nm and an emission wavelength of 617 nm. Images were taken at ×63.
HDAC4 antibody and confocal detection
PC3 cells were treated with drug at the final concentration of 1 µmol/L for 24 hours. The cells were then washed with 1 × PBS and fixed with 4% paraformaldehye for 15 minutes. Nonspecific binding was blocked with 10% normal goat serum in PBS containing 0.2% tween-20 (Sigma Aldrich P7949) for 1 hour. Cells were treated with mouse monoclonal anti-HDAC4 antibody (Sigma Aldrich H0163) 1:200 dilution in 5% goat serum in PBS for 1 hour at room temperature. Fluorescent rabbit anti-mouse monoclonal antibodies (Alexa fluor 488 from Invitrogen A11001) were applied to the cells for 30 minutes. After washing, Vecta shield mounting medium for fluorescence with DIPA (Vector laboratory H-1200) was loaded on the cells and covered with cover glass for imaging.
Western blot analysis
Cells were treated 24 hours after seeding with compound (dissolved in DMSO) at the concentration of (0.1, 1, 5, 10, and 50 µmol/L), and an equal amount of DMSO was added to the control groups. After incubation at 37°C, the cell culture media was removed and washed once with fresh media, then HDAC compounds were added to the cell culture. The cell culture was kept at 37°C for another 24 hours, then harvested. The cells were harvested by washing 2 times with PBS and lysed with radioimmunoprecipitation assay buffer. The protein concentrations were measured by Bradford protein assay method. Equal amounts of proteins were mounted on the 4% to 20% SDS-PAGE gel, with antibody against acetylated α-tubulin (Sigma) and acetylated histone H3 (Cell Signaling). For Western blot analysis, proteins were separated in 4% to 20% SDS-PAGE gels and blotted onto ImmobilonP membrane (Millipore). ECL detection (Amersham) was done according to manufacturer’s instructions.
Results
Rational drug design and molecular modeling
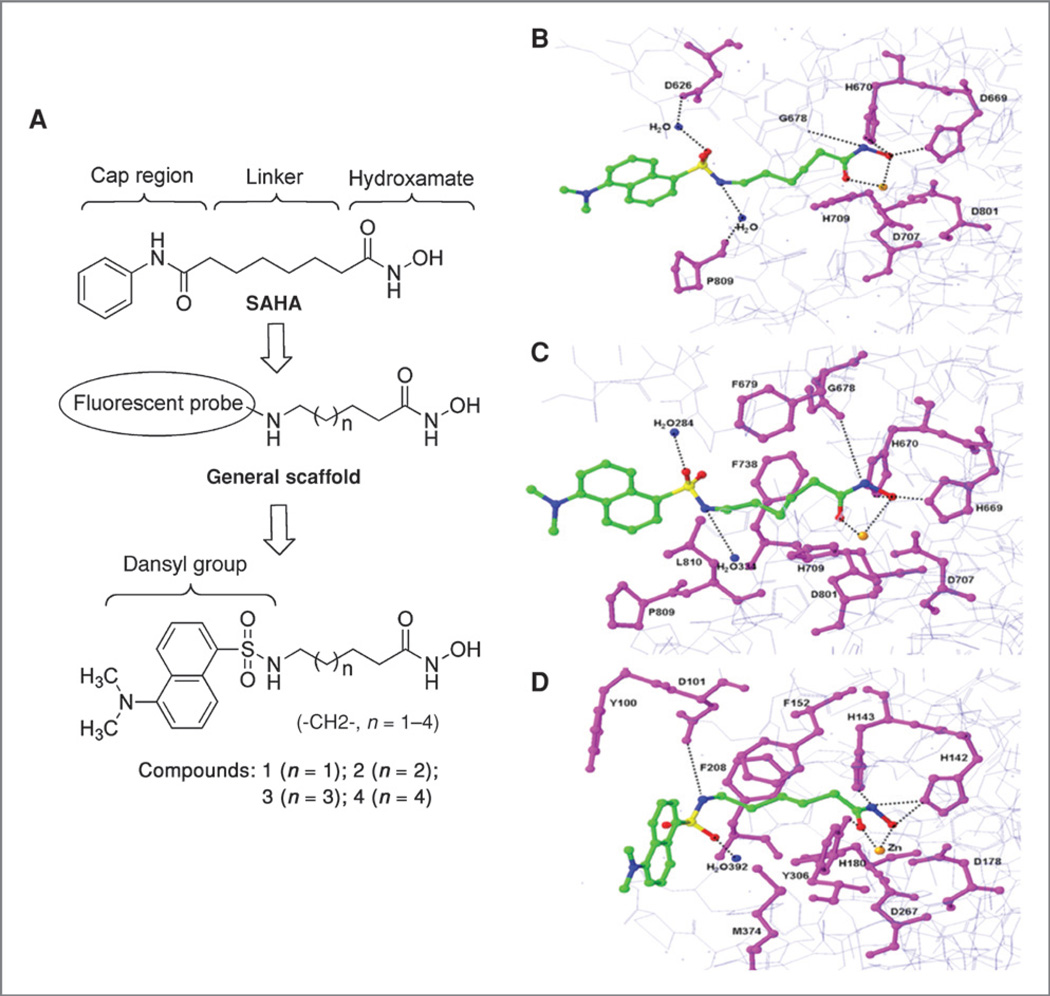
We developed homology models to represent HDAC isomers based on the observation that each HDAC is unique in the CAP region. Substitution of bulky cap groups potentially offers increased selectivity and potency, and the site is capable of accommodating a large fluorescent probe. Docking simulations of compounds 1 to 4 (Fig. 1A) against HDAC isomers led to comparisons of root mean SDs and molecular dynamics simulations on HDACs 6, 7, and 8. The results of 100 ps molecular dynamic simulations of compound 2 are illustrated in Fig. 1B–D. The planar ring of the dansyl moiety contributes to affinity by preferentially occupying the relatively planar and hydrophobic portion of the CAP region (Fig. 1B–D).
Figure 1.
A, design of fluorescent hydroxamic acid HDAC inhibitors. B–D, proposed binding model of 2 (green) in complex with HDAC6, 7, and 8. X-ray structures of HDAC7 and HDAC8 were used, whereas HDAC6 is a homology model based on HDAC7. Highlighted residues that make critical interactions with compound 2 are represented by the stick model (magenta). Hydrogen bonds are indicated by dotted line (black), zinc shown as orange, and water shown as blue.
Synthesis of compound 2 and analogues
The strategy for synthesis of fluorescent molecule 2 and analogues containing various length linkers is outlined in Supplementary Fig. S1. In general, dansyl chloride was reacted with the appropriate amino carboxylic acid to yield the dansylated derivative acids 5 to 8 in high yield. By using an EDCI-HOBT combined condensation, acids 5 to 8 were activated and condensed with benzyloxyamine to yield amides 9 to 12. Hydrogenation of 9 to 12 provided the final hydroxamic acid products 1 to 4 with yields more than 90%.
Screening for HDAC inhibitory activity
In vitro pan-HDAC activity inhibition assays were done as the initial screen to assess overall compound potencies. Compounds 2 and 3 are effective pan-HDAC inhibitors with IC50 values of 125 and 116 nmol/L, respectively, as compared with compounds 1 and 4 (Table 1). The inhibitory activities of these compounds were linker-length dependent. The 4- and 7-carbon spacers (compounds 1 and 4) were less active than were the 5- and 6-carbon linkers (compound 2 and 3), which allowed appropriate positioning of the hydroxyamic acid and the dansyl fluorophore within the hydrophobic cavity (Fig. 1B–D).
Table 1.
Physicochemical properties and pan-HDAC inhibition activities
| Compound | Carbon linker (n) |
HDAC IC50 [nmol/L] |
PSA | MW | CLogP | MV(Å)3 | Excitation (nm) |
Emission (nm) |
|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 287 | 213.7 | 365.45 | 1.19 | 1,037.4 | 320 | 500 |
| 2 | 2 | 125 | 218.4 | 379.48 | 1.72 | 1,067.5 | 320 | 500 |
| 3 | 3 | 116 | 218.7 | 393.50 | 2.25 | 1,104.2 | 320 | 500 |
| 4 | 4 | 208 | 216.3 | 407.53 | 2.78 | 1,194.5 | 320 | 500 |
| SAHA | 80 | 180.1 | 264.32 | 0.99 | 868.7 |
NOTE: Compound structures are shown in Fig. 1.
Abbreviations: PSA, polar surface area; MW, molecular weight; CLogP, calculated log of the partition coefficient; MV, molecular volume; Excitation, excitation wavelength; Emission, emission wavelength.
HDAC inhibitory potency of compound 2 was also maintained by substituting a sulfonamide in the linker unit in place of a carbonylamide. The dansyl moiety is exposed in the CAP region and forms vertical pi–pi stacking interactions with surrounding hydrophobic residues. Compound 3 contains a 6-carbon spacer flexibly positioned into the hydrophobic cavity with hydrogen bonding similar to that of compound 2 (structure figure not shown), however, the 7-carbon linker in compound 4 does not position the dansyl group optimally (figure not shown). Although the 4-carbon spacer of compound 1 fits well into the hydrophobic site, the lack of H-bond formation of the amide group (contained in the dansyl moiety) with the surface residues results in low affinity (Table 1). These data show that compounds containing dansyl groups in the cap regions offer effective inhibition of HDACs.
The compounds fall well within the rules proposed by Lipinski (15), and we also note that the polar surface areas of the compounds are all greater than 150 Å2. Measurements of the excitation and emission spectra for compounds 1 to 4 document the fluorescence properties of the HDAC inhibitors (Table 1).
HDAC isomer specificity
Observations of nonoverlapping functions for HDAC isomers present an opportunity for achieving therapeutic specificities. Compounds 2 and 3 were evaluated for inhibitory effects on recombinant proteins available for 10 of the class I and II HDAC isomers. As summarized in Table 2, compounds 2 and 3 show increased inhibition of class II HDAC isomers. Of note, HDAC6 is a microtubule-associated protein that can deacetylate α-tubulin in vitro. Also, HDAC6 has been reported to interact with β-tubulin in vitro (16). This dual deacetylase function of HDAC6 may offer a research tool to study cellular function of HDACs. Because HDAC6 localizes predominantly in the cytoplasm and deacetylates tubulin, we considered it as functionally distinct from other known HDACs and focused on substrates other than histones.
Table 2.
HDAC isomer inhibition activity
| HDAC isom er IC50 (µmol/L)a | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Compound | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 10 | 11 |
| 2 | 0.95 | 1.38 | 1.12 | 0.33 | 0.40 | 0.13 | 2.56 | 3.98 | 0.42 | 0.48 |
| 3 | 1.29 | 7.40 | 4.89 | 0.67 | 0.37 | 0.08 | 6.40 | 2.58 | 0.42 | 0.52 |
| SAHA | 0.22 | 0.56 | 1.79 | 0.64 | 0.13 | 0.027 | 0.99 | 2.74 | 0.11 | 0.082 |
HDAC class I (1, 2, 3, and 8); class II (4, 5, 6, 7, 9, and 10); class IV (11).
Figure 1 shows the molecular model of compound 2 in complex with HDAC6 (Fig. 1B), HDAC 7 (Fig. 1C), and HDAC8 (Fig. 1D). Interactions of the dansyl moiety within the CAP regions of HDAC isomers provide a potential basis for selectivity. Comparison of HDAC isomers 6 and 8 reveals that the shape of the HDAC6-binding cavity differs significantly. This data along with differences in HDAC isomer inhibition further support that there may be significant conformational or structural dissimilarity in class I and class II HDAC isomers. Furthermore, electrostatic and steric surface interactions around the ligand-binding channel show differences between class I and class II HDACs.
Cell growth inhibition and cell cycle distributions
As promising targets for cancer treatment, HDAC inhibitors have shown significant antiproliferative and apoptotic activities on various cancer cells, including prostate cancer (17–19). HDAC inhibitors are capable of inducing differentiation and/or apoptosis of prostate cancer cells. Therefore, we evaluated compound 2 for growth inhibition of 3 human prostate cancer cell lines including PC-3, C4-2, and LNCaP. Compound 2 inhibited the growth of all 3 prostate cancer cell lines with GI50s for PC3 (1.54 ± 0.27 µmol/L), C4–2 (1.91 ± 0.15 µmol/L), and LNCaP (1.30 ± 0.41 µmol/L). Treatment with GI50 doses of compound 2 resulted in an increase of the percentage of cells in G1 as compared with S or G2 (Table 3). This effect was observed in both androgen-dependent and -independent prostate cancer cells. We also evaluated compound 2 against additional human cancer cell lines MCF7 and A549 (Table 3). The effects of our compound on cell cycle distributions were similar to that observed with SAHA.
Table 3.
Effect of compound 2 on cell cycle distribution
| Compound | Cell line | Cell cycle distribution (%) |
||
|---|---|---|---|---|
| G1 | S | G2 | ||
| Control | LNCaP | 72.50 | 8.95 | 8.55 |
| 2 | LNCaP | 85.23 | 9.18 | 5.59 |
| Control | PC3 | 47.29 | 21.15 | 31.56 |
| 2 | PC3 | 64.35 | 19.91 | 15.74 |
| SAHA | PC3 | 72.28 | 14.33 | 13.39 |
| Control | A549 | 79.27 | 9.81 | 10.93 |
| 2 | A549 | 82.63 | 2.03 | 15.34 |
| SAHA | A549 | 88.58 | 2.00 | 9.41 |
| Control | MCF-7 | 62.03 | 29.73 | 8.24 |
| 2 | MCF-7 | 75.76 | 12.94 | 11.30 |
| SAHA | MCF-7 | 77.43 | 6.68 | 15.89 |
NOTE: GI50s of SAHA at 1.04 µmol/L for PC3 and the compound 2 at 1.54 µmol/L for PC3 and 1.3 µmol/L for LNCaP were used. A549 cells were treated with 10 µmol/L of compound 2 and SAHA. No obvious change at 1 µmol/L concentration was observed. MCF-7 cells were treated with 1 µmol/L of compound 2 and SAHA.
Imaging compound 2 in human prostate cancer cells
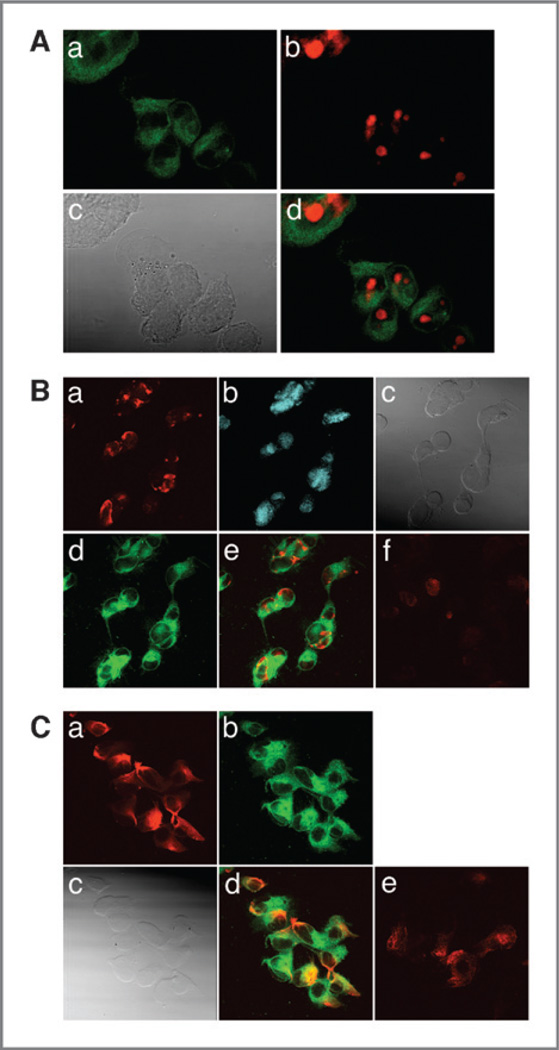
By utilizing the inherent fluorescent properties of our compound, we imaged compound 2 in human prostate cancer cells with confocal miscroscopy. The addition of 20 µmol/L of compound 2 to PC3 (Supplementary Fig. S2A–C) and DU145 (Supplementary Fig. S2D–F) cells showed intracellular cytoplasmic delivery. The images shown here were obtained 60 minutes after cell treatment. Similar results were obtained after 8 and 16 hours, confirming that compound 2 localized to the cytoplasm. Colocalization experiments with propidium iodide revealed that compound 2 does not localize to nuclei (Fig. 2A).
Figure 2.
A, colocalization of fluorescent compound 2 in PC-3 cells. Images were taken at 695 nm with a multiphoton laser. Cells were treated with 20 µmol/L of compound 2 and images were obtained 60 minutes following drug exposure. a, cytoplasm contains fluorescent compound 2 (green); b, nucleus is stained with propidium iodide (red); c, differential interference contrast image (gray); d, merged image of a, b, and c. B, compound 2 effects on nuclear histone acetylation in A549 cells. Images were taken at 695 nm with a multiphoton laser. a, acetylated histone (red); b, propidium iodide stain of the nucleus; c, differential interference contrast image; d, 20 µmol/L of compound 2 (green) shown in cytoplasm; e, fused image of a, b, c and d; f, control acetylation. C, effects of compound 2 on tubulin acetylation in the cytoplasm of A549 cells. Images were taken at 695 nm with a multiphoton laser. a, acetylated tubulin (red). b, 20 µmol/L of fluorescent compound 2 (green); c, differential interference contrast image; d, merged image of a, b, and c; e, control acetylated tubulin (without compound 2).
Imaging the effects of compound 2 on histone acetylation
Next, we determined the effects of compound 2 on histone acetylation in A549 cells. A549 cells were treated with 20 µmol/L of compound 2, allowing comparisons to previously published data (20) and evaluation of this novel compound in other cancer cell types. Figure 2B shows that compound 2 increases acetylation of nuclear histones, a marker of the functional effect on the enzyme, as compared with controls, which was not shown clearly in the literature (Fig. 2B, f; ref. 20). Interestingly, compound 2 localized to the cytoplasm (Fig. 2B, d) and not in the nucleus, prompting a reevaluation of the accepted mechanisms of HDAC inhibitor action in the nucleus (21, 22).
Imaging the effects of compound 2 on nonhistone protein
Class II HDACs shuttle between the nuclei and cytoplasmic compartments of cells and modify the acetylation status of tubulin residing in the cytoplasm. Compound 2 also caused an increase in α-tubulin acetylation (Fig. 2C) in the cytoplasms of A549 cells as compared with control (Fig. 2C, e). As shown in experiments using other cell lines, compound 2 fluoresced in the cytoplasms of these cells as well (Fig. 2C, b).
Mechanistic studies on HDAC4 interactions with compound 2
HDAC isomers show nonredundant functions. HDAC6 is a microtubule-associated protein that deacetylates tubulin in vitro. The expression of HDAC6 has been reported exclusively in the cytoplasm (23). HDAC10 was found to be the closest relative of HDAC6, but it has an incomplete second HDAC domain (24–26). Other class II HDACs, for example HDAC4 and HDAC5, have been found to shuttle between the nucleus and the cytoplasm (27, 28), whereas HDAC7 shuttling is not certain. This correlates to the observations that compound 2 is potent in inhibiting HDAC 4, 5, 6, and 10 and also has shown activity against HDAC11, which combines properties of both class I and class II HDACs (Table 2). We interpret these data to show that compound 2 affects the functions of the nuclear HDAC enzymes without entering the nucleus.
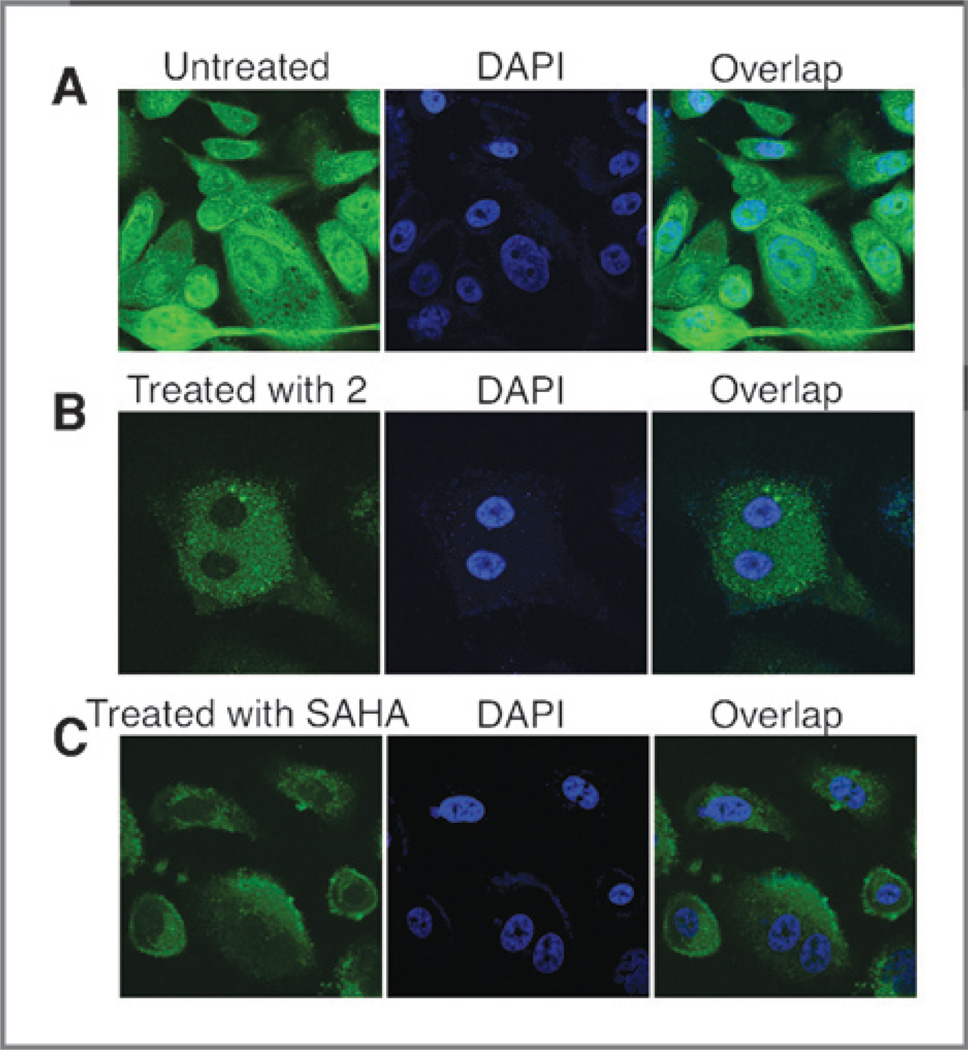
Compound 2 fluorescence was detected in the cytoplasm; therefore, we sought to evaluate its interactions with HDACs capable of shuttling from the nucleus into the cytoplasm. HDAC4 was selected as the model HDAC for these studies given its known cytoplasmic nuclear shuttling (29) and the potency (IC50, Table 2) of compound 2 against HDAC4. By using an antibody to HDAC4, we evaluated HDAC4 distribution in PC3 cells with/without inhibitor treatment (Fig. 3A–C). Confocal microscopy shows the expected cytoplasmic and nuclear distributions of HDAC4 (Fig. 3A). Treatment of PC3 cells with 1 µmol/L of compound 2 resulted in a significant reduction of HDAC4 in the nucleus. The 4′,6-diamidino-2-phenylindole staining of the nucleus confirmed depletion of HDAC4 (Fig. 3B). Interestingly, in control experiments, SAHA also depleted the nucleus of HDAC4 (Fig. 3C). As a control, we evaluated by Western analysis of 5 HDAC isomers including HDAC1 and 2 and found no change in protein concentration up to 50 µmol/L dose (See Supplementary Fig. S3). In addition, PC3 cells treated with compound 2 and immunostained for HDAC1 continued to show HDAC1 in the nucleus (See Supplementary Fig. S4). These observations show the usefulness of the new HDAC ligand and point to potential new interpretations of mechanisms for inhibition of nuclear HDAC-mediated deacetylation.
Figure 3.
HDAC inhibitor trap HDAC4 in the cytoplasm of human prostate cancer cells. A, untreated PC3 cells. B, PC3 cells treated with 1 µmol/L of compound 2 for 24 hours and detected for HDAC4. C, PC3 cells treated with 1 µmol/L SAHA for 24 hours and detected for HDAC4. Images were taken at 695 nm with a multiphoton laser.
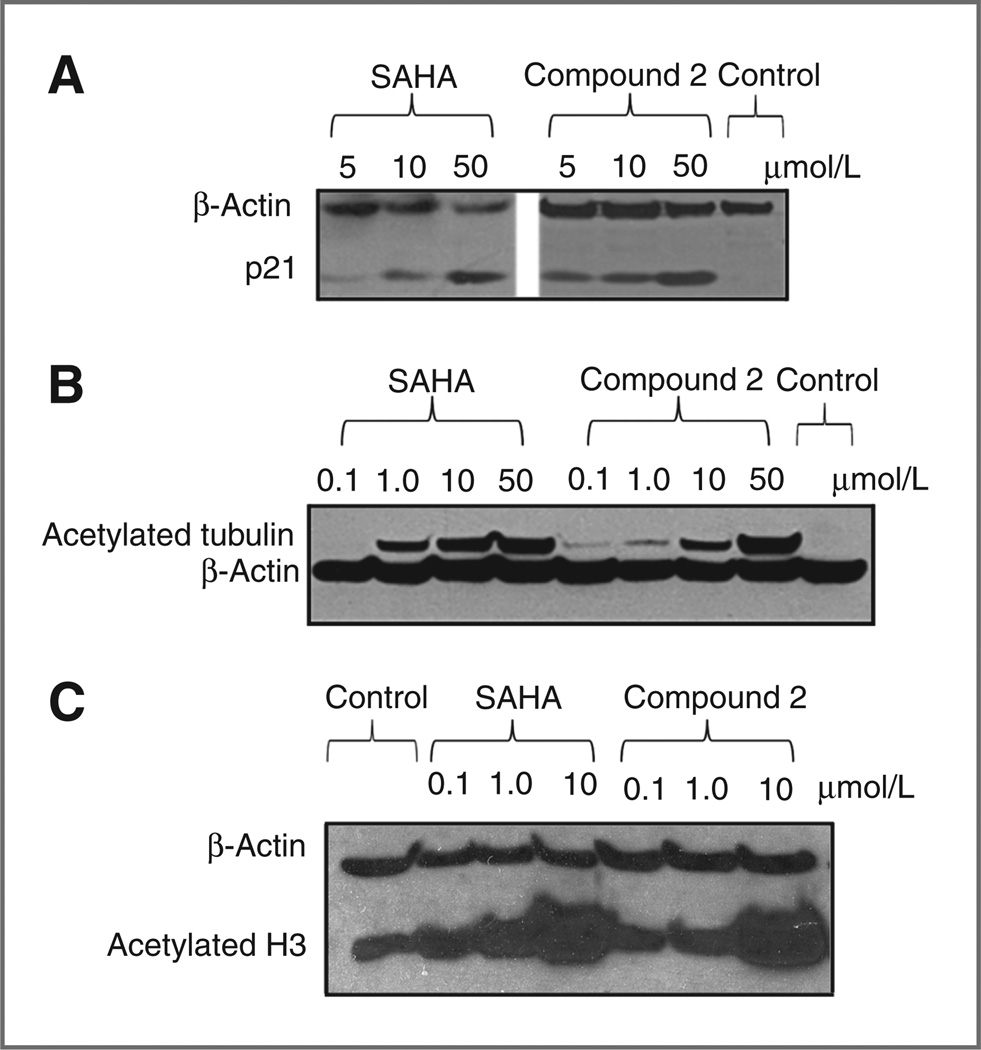
The transcription of p21 is regulated through acetylation of histone. Acetylation of histones results in increased p21 transcription. Hence, the expression levels of p21 reflect the inhibitory effects on HDACs by these compounds. PC3 cells do not express p53, resulting in decreased p21 levels (30, 31). T24 cells (human bladder cancer cells) were used to check the expression levels of p21 following treatment with compound 2. Exposure of T24 cells to 5 µmol/L of compound 2 resulted in upregulated expression of p21 (Fig. 4A).
Figure 4.
A, p21 expression in PC3 cells. B, acetylated tubulin expression in T24 cell lines. C, acetylated histone 3 (H3) in PC3 cells when treated with compound 2.
p21 binds to and inhibits cyclin-CDK2, resulting in G1 arrest (32). Compound 2 caused G1 arrest in PC3 and LNCaP cells (Table 3). Because PC3 cells do not express p53, these data suggest the presence of an alternative pathway to G1 arrest.
HDACs play important roles in regulating other cellular activities. Class II HDAC shuttle between the nuclei and cytoplasm and modify the acetylation status of tubulin residing in the cytoplasm. When PC3 cells were treated with compound 2 or with SAHA, the acetylation levels of tubulin (Fig. 4B) and H3 (Fig. 4C) were increased. This further supports our findings with the HDAC isomer inhibition assay (Table 2). Compound 2 showed potent inhibition of class II HDACs, especially HDAC6.
HDAC6 is known to affect protein stability (33), a potentially increasing degradation of class I HDACs. We evaluated 5 HDAC isomers including HDAC1, 2, 3, 4, and 5 and observed no changes in protein levels up to 50 µmol/L exposure (Supplementary Fig. S3).
Discussion
In this study, we have designed and synthesized novel fluorescent HDAC inhibitors. We report compounds 2 and 3, HDAC class II selective hydroxamate inhibitors, especially targeting HDAC4 and HDAC6. Compound 2 not only exhibited general properties of hydroxamate HDAC inhibitors, inhibiting cancer cell growth and causing G1 arrest but was also found exclusively in the cytoplasm of cells. Interestingly, treatment of cells with compound 2 resulted in increased nuclear acetylation. We attribute this observation to HDAC4 sequestration, shifting the balance of histone acetylation to a more acetylated state. This compound also inhibited tubulin deacetylation in the cytoplasm of human lung cancer cells, supporting its action on HDAC6. These activities are promising for developing the compounds as potential cancer therapies.
Our results showing the localization of a hydroxamate HDAC inhibitor in the cytoplasm may offer a new paradigm for understanding how HDAC inhibitors affect nuclear function of HDACs. This mechanism may be related to HDAC trapping of shuttled proteins. Cytoplasmic localization of compound 2 further leads to speculation on (i) the effects of compound 2 on nuclear translocation of HDAC proteins, (ii) potential inhibitory effects on the process of mRNA nuclear exportation, (iii) cytoplasmic HDAC degradation, and (iv) effects of compound 2 on protein folding or interactions with nuclear translocation or chaperone proteins. Also, restriction of HDAC4 in the cytoplasm may result in disruption of nuclear complexes that include HDAC4.
Taken together, these observations suggest that compound 2 leads to modified expression and functions of molecules important for cellular responses by confining HDACs to the cytoplasmic compartments. This finding provides a valuable tool for further studies to understand the mechanism of the action of HDAC inhibitors. Further studies of the involved isomers and mechanisms are needed to provide a comprehensive mechanistic understanding.
Acknowledgments
We thank Advanced Biomedical Computing Center of the National Cancer Institute, Frederick, MD, for allocation of computing time and staff support.
Grant Support
This work was financially supported by the Georgetown University Drug Discovery Program and the Shared Resources at the Lombardi Comprehensive Cancer Center (Grant NIH/NCI # P30 CA51008).
Footnotes
Supplementary material for this article is available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
A patent application has been filed by Georgetown University on the behalf of the inventors that are listed as authors in this article.
References
- 1.Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 2.Laherty CD, Yang WM, Sun JM, Davie JR, Seto E, Eisenman RN. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 3.Emiliani S, Fischle W, Van Lint C, Al-Abed Y, Verdin E. Characterization of a human RPD3 ortholog, HDAC3. Proc Natl Acad Sci U S A. 1998;95:2795–2800. doi: 10.1073/pnas.95.6.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu E, Chen Z, Fredrickson T, Zhu Y, Kirkpatrick R, Zhang GF, et al. Cloning and characterization of a novel human class I histone deacetylase that functions as a transcription repressor. J Biol Chem. 2000;275:15254–15264. doi: 10.1074/jbc.M908988199. [DOI] [PubMed] [Google Scholar]
- 5.Grozinger CM, Hassig CA, Schreiber SL. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc Natl Acad Sci U S A. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 7.De Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Ashburner BP, Baldwin AS, Westerheide SD., Jr. The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) core-pressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol Cell Biol. 2001;21:7065–7077. doi: 10.1128/MCB.21.20.7065-7077.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blagosklonny MV, Robey R, Sackett DL, Du L, Traganos F, Darzyn-kiewicz Z, et al. Histone deacetylase inhibitors all induce p21 but differentially cause tubulin acetylation, mitotic arrest, and cytotoxicity. Mol Cancer Ther. 2002;1:937–941. [PubMed] [Google Scholar]
- 11.Naryzhny SN, Lee H. The post-translational modifications of proliferating cell nuclear antigen: acetylation, not phosphorylation, plays an important role in the regulation of its function. J Biol Chem. 2004;279:20194–20199. doi: 10.1074/jbc.M312850200. [DOI] [PubMed] [Google Scholar]
- 12.Varshochi R, Halim F, Sunters A, Alao JP, Madureira PA, Hart SM, et al. ICI182,780 induces p21Waf1 gene transcription through releasing histone deacetylase 1 and estrogen receptor alpha from Sp1 sites to induce cell cycle arrest in MCF-7 breast cancer cell line. J Biol Chem. 2005;280:3185–3196. doi: 10.1074/jbc.M408063200. [DOI] [PubMed] [Google Scholar]
- 13.Jung M, Kozikowski A, Dritschilo A. Rational design and development of radiation-sensitizing histone deacetylase inhibitors. Chem Biodi-vers. 2005;2:1452–1461. doi: 10.1002/cbdv.200590118. [DOI] [PubMed] [Google Scholar]
- 14.Chen B, Petukhov PA, Jung M, Velena A, Eliseeva E, Dritschilo A, et al. Chemistry and biology of mercaptoacetamides as novel histone deacetylase inhibitors. Bioorg Med Chem Lett. 2005;15:1389–1392. doi: 10.1016/j.bmcl.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Li N, Caron C, Matthias G, Hess D, Khochbin S, et al. HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo . EMBO J. 2003;22:1168–1179. doi: 10.1093/emboj/cdg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy S, Packman K, Jeffrey R, Tenniswood M. Histone deacetylase inhibitors differentially stabilize acetylated p53 and induce cell cycle arrest or apoptosis in prostate cancer cells. Cell Death Differ. 2005;12:482–491. doi: 10.1038/sj.cdd.4401581. [DOI] [PubMed] [Google Scholar]
- 18.Butler LM, Agus DB, Scher HI, Higgins B, Rose A, Cordon-Cardo C, et al. Suberoylanilide hydroxamic acid, an inhibitor of histone deacetylase, suppresses the growth of prostate cancer cells in vitro and in vivo . Cancer Res. 2000;60:5165–5170. [PubMed] [Google Scholar]
- 19.Kuefer R, Hofer MD, Altug V, Zorn C, Genze F, Kunzi-Rapp K, et al. Sodium butyrate and tributyrin induce in vivo growth inhibition and apoptosis in human prostate cancer. Br J Cancer. 2004;90:535–541. doi: 10.1038/sj.bjc.6601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong JC, Hong R, Schreiber SL. Structural biasing elements for in-cell histone deacetylase paralog selectivity. J Am Chem Soc. 2003;125:5586–5587. doi: 10.1021/ja0341440. [DOI] [PubMed] [Google Scholar]
- 21.Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26:5541–5552. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- 22.Gui CY, Ngo L, Xu WS, Richon VM, Marks PA. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proc Natl Acad Sci U S A. 2004;101:1241–1246. doi: 10.1073/pnas.0307708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verdel A, Khochbin S. Identification of a new family of higher eukaryotic histone deacetylases. Coordinate expression of differentiation-dependent chromatin modifiers. J Biol Chem. 1999;274:2440–2445. doi: 10.1074/jbc.274.4.2440. [DOI] [PubMed] [Google Scholar]
- 24.Guardiola AR, Yao TP. Molecular cloning and characterization of a novel histone deacetylase HDAC10. J Biol Chem. 2002;277:3350–3356. doi: 10.1074/jbc.M109861200. [DOI] [PubMed] [Google Scholar]
- 25.Kao HY, Lee CH, Komarov A, Han CC, Evans RM. Isolation and characterization of mammalian HDAC10, a novel histone deacetylase. J Biol Chem. 2002;277:187–193. doi: 10.1074/jbc.M108931200. [DOI] [PubMed] [Google Scholar]
- 26.Tong JJ, Liu J, Bertos NR, Yang XJ. Identification of HDAC10, a novel class II human histone deacetylase containing a leucine-rich domain. Nucleic Acids Res. 2002;30:1114–1123. doi: 10.1093/nar/30.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKinsey TA, Zhang CL, Olson EN. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc Natl Acad Sci U S A. 2000;97:14400–14405. doi: 10.1073/pnas.260501497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verdel A, Curtet S, Brocard MP, Rousseaux S, Lemercier C, Yoshida M, et al. Active maintenance of mHDA2/mHDAC6 histone-sdeacetylase in the cytoplasm. Curr Biol. 2000;10:747–749. doi: 10.1016/s0960-9822(00)00542-x. [DOI] [PubMed] [Google Scholar]
- 29.Zhao X, Ito A, Kane CD, Liao TS, Bolger TA, Lemrow SM, et al. The modular nature of histone deacetylase HDAC4 confers phosphorylation-dependent intracellular trafficking. J Biol Chem. 2001;276:35042–35048. doi: 10.1074/jbc.M105086200. [DOI] [PubMed] [Google Scholar]
- 30.Scott SL, Earle JD, Gumerlock PH. Functional p53 increases prostate cancer cell survival after exposure to fractionated doses of ionizing radiation. Cancer Res. 2003;63:7190–7196. [PubMed] [Google Scholar]
- 31.Zhang Z, Li M, Wang H, Agrawal S, Zhang R. Antisense therapy targeting MDM2 oncogene in prostate cancer: Effects on proliferation, apoptosis, multiple gene expression, and chemotherapy. Proc Natl Acad Sci U S A. 2003;100:11636–11641. doi: 10.1073/pnas.1934692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bao GC, Wang JG, Jong A. Increased p21 expression and complex formation with cyclin E/CDK2 in retinoid-induced pre-B lymphoma cell apoptosis. FEBS Lett. 2006;580:3687–3693. doi: 10.1016/j.febslet.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 33.Sadoul K, Boyault C, Pabion M, Khochbin S. Regulation of protein turnover by acetyltransferases and deacetylases. Biochimie. 2008;90:306–312. doi: 10.1016/j.biochi.2007.06.009. [DOI] [PubMed] [Google Scholar]