Fig. 3.
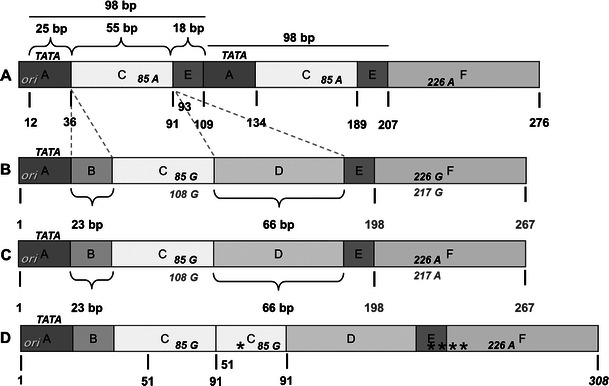
PML-associated variant Mad-1 (A), archetype CY (B) and JCV NCCR structures found in samples (C, D). The nucleotide sequences are shown from the core of the origin of DNA replication (ori) to the start site of the late leader protein, Agno protein. In A, the nucleotide numbering is based on PML-associated variant Mad-1 NCCR sequence and numbers are indicated in bold and black. In B, the nucleotide numbering of the archetype sequence of Japanese strain CY, isolated by Yogo et al. (1991) is reported in bold and grey. In C, the archetype variant sequence found in CSF specimens collected at t0 and t3, in plasma at t1 and t3 and in PBMC at t0, t1 and t3 is reported. In D, the rearranged JCV NCCR sequence of 308 bp in length, obtained from the CSF samples collected at t1and t2, is showed. Asterisks represent single nucleotide point mutations or deletions. The TATA box is presented by TATA. Italicised capital letters indicate nucleotides. Mad-1 NCCR contains an adenine at positions 85 and 183, compared with archetype CY NCCR, which contains guanine at these positions. Boxes division from A to F is also shown in capital letters. The sequence analysis was performed directly on DNA template previously amplified by nested-PCR, using two pairs of primers that anneal to the invariant regions flanking the NCCR of JCV. Primers BKTT1 (5′-AAG GTC CAT GAG CTC CAT GGA TTC TTC C-3′) and BKTT2 (5′-CTA GGT CCC CCA AAA GTG CTA GAGCAG C-3′) amplified a 724-bp DNA fragment in JCV (Mad-1). The second pair, JC1 (5′-CCT CCA CGC CCT TAC TAC TTC TGA G-3′) and JC2 (5′-AGC CTG GTG ACA AGC CAA AAC AGC TCT-3′), amplified a portion of the first round PCR product, generating a fragment of 308 bp (Pietropaolo et al. 2003)
