Figure 7.
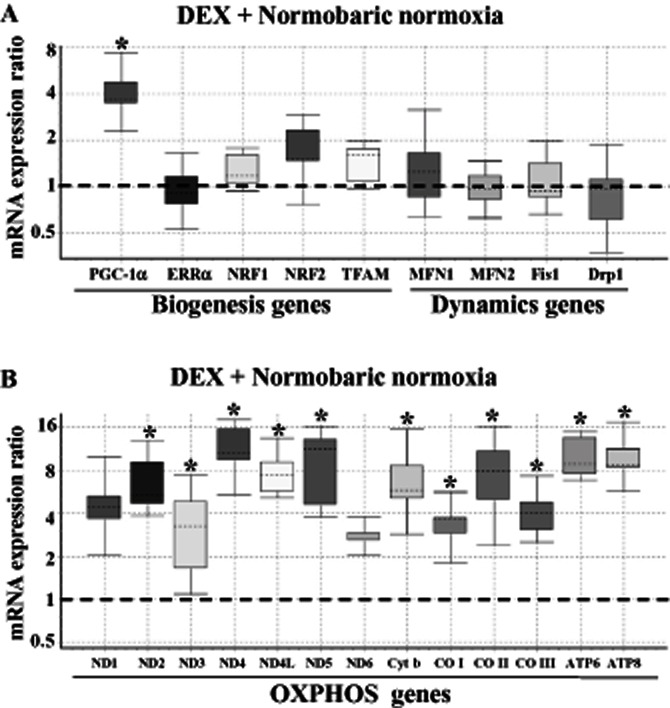
Effect of DEX (or HH adaptation) on the relative expression of mitochondrial biogenesis/dynamics genes (A) and mtDNA-encoded OXPHOS genes (B). The relative expression of genes in the lung tissue derived from DEX pretreated rats under NN conditions was measured using QRT-PCR as detailed under Materials and Methods. The Box and whisker plot reports the mRNA expression ratio on the y-axis of different genes denoted on the x-axis. The expression ratio of DEX pretreated rat lung was calculated relative to the control group (i.e. without DEX treatment) maintained in NN conditions (fixed at 1) using β-actin gene expression for normalization. Note that DEX selectively increased (∼3.5-fold) the PGC-1α mRNA transcript. However, other genes involved in mitochondrial biogenesis/dynamics were maintained at NN levels only. Also, DEX treatment augmented (∼2- to 12-fold) the expression of mtDNA-encoded OXPHOS genes, which is highly significant (*P(H1) < 0.05 in comparison with the control group; n = 3 per group). The name of the genes as abbreviated in x-axis is given in expansion under abbreviations section.
