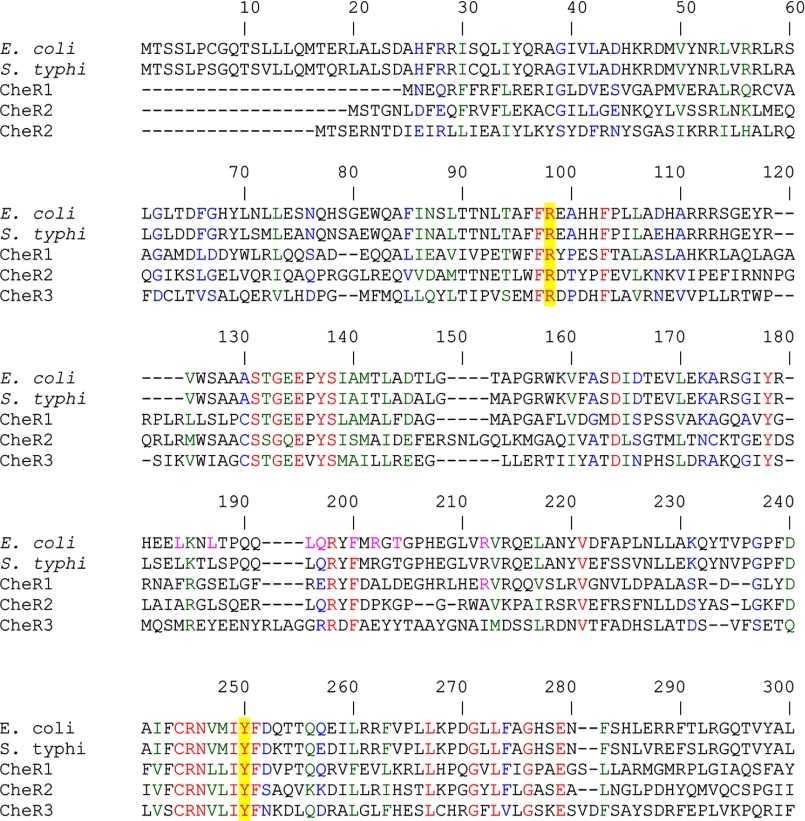
FIGURE 3.
Segment of sequence alignment of the 3 CheR paralogues from P. putida KT2440 with the CheR sequences form E. coli (P07364) and S. typhimurium (P07801). Sequences were aligned using ClustalW of the Network Protein Sequence Analysis. The GONNET protein weight matrix was used with a gap opening penalty of 10 and gap extension penalty of 1. Amino acids in red indicate identity, in blue indicate high similarity, and in green indicate low similarity. The amino acids that are involved in catalysis are shaded in yellow. The amino acids corresponding to the amino acids Arg-98 and Tyr-235 in S. typhimurium CheR are shaded in yellow.