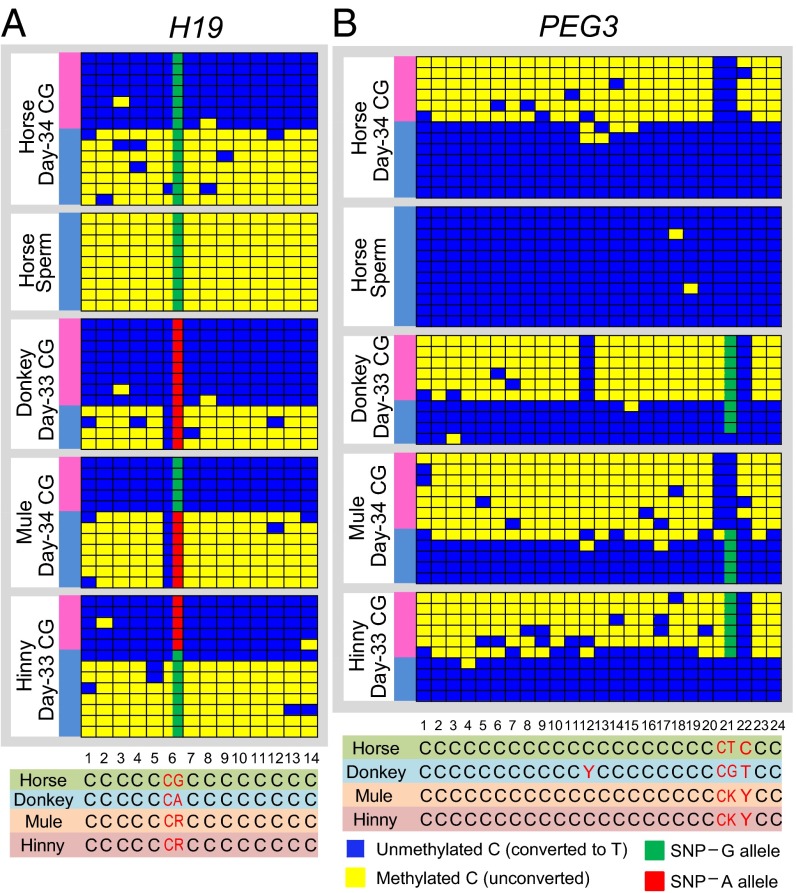
Fig. 3.
Strand-specific methylation of CpG islands of equine imprinted genes. Bisulfite sequencing of the DMRs in H19 and PEG3 shows allele-specific differential methylation corresponding to the allelic expression bias. Parental allele-specific methylation in mule and hinny is inferred from differences in single-nucleotide sequences in horse and donkey. Yellow boxes depict methylated CpGs, and blue boxes depict unmethylated CpGs. Each panel shows multiple CpG sites at the DMR for the corresponding gene. (A) H19 is 100% methylated in sperm, consistent with paternal silencing. The H19 DMR is differentially methylated in day 33–34 chorionic girdle (CG) samples from horse, donkey, mule, and hinny, with paternal methylation. Note the concordance between the SNP allelic state and the methylation state in mule and hinny. (B) PEG3 is unmethylated in sperm, consistent with paternal expression. The PEG3 DMR is differentially methylated in day 33–34 chorionic girdle samples from horse, donkey, mule, and hinny, with maternal methylation.