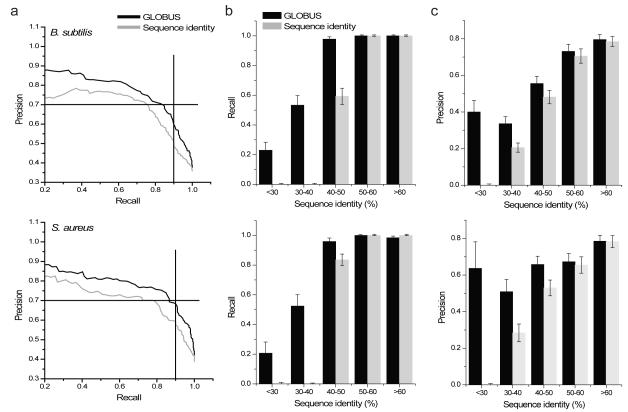
Figure 2. GLOBUS precision-recall performance.
Using available metabolic models (iBsu110333 for B. subtilis and iSB61935 for S. aureus) we compared predictions by GLOBUS to predictions made using sequence homology; predictions for B. subtilis are on the top, and predictions for S. aureus are on the bottom. (a) Precision–recall curves for GLOBUS (black lines) were calculated by ranking genes using assignment probabilities. Precision-recall curves for homology (red lines) were calculated by ranking genes using sequence identity. (b) Recall of known metabolic genes (at 70% precision) as a function of sequence identity to the closest enzymes from other species with the annotated functions. (c) Prediction precision (at 90% recall) for known metabolic genes as a function of sequence identity to the closest enzymes from other species with the annotated functions. In the figure error bars represent the S.E.M,