Figure 2.
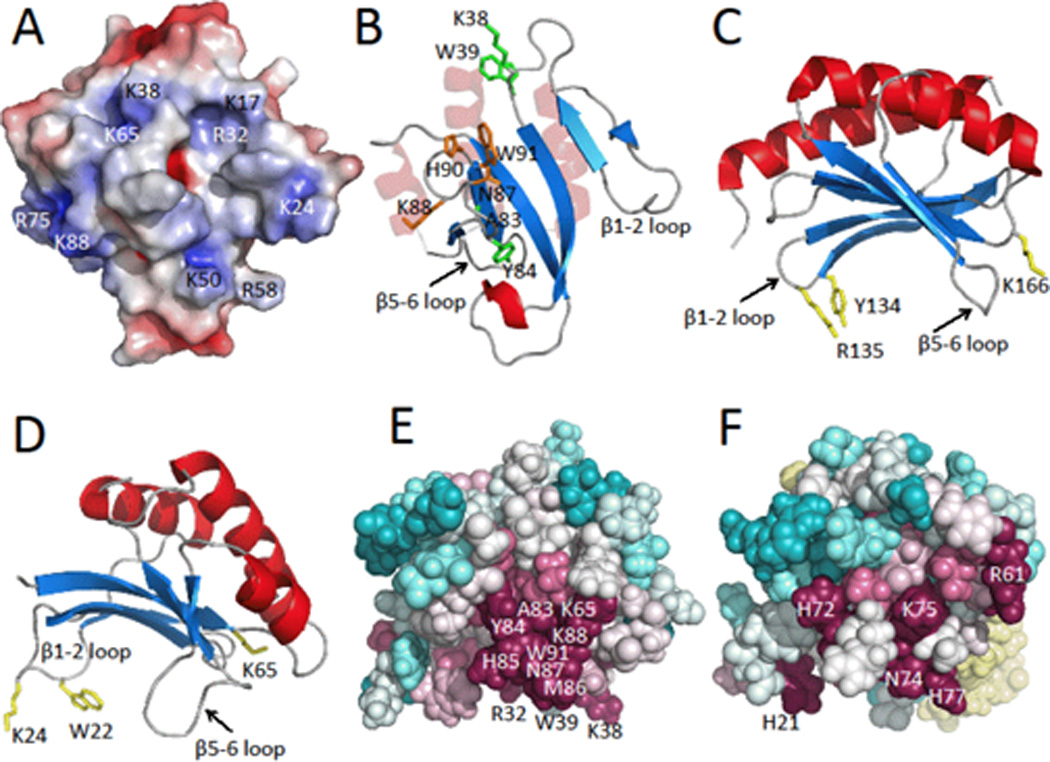
A) Electrostatic surface potential map of the lowest energy NMR conformer of Pspto_3016 generated with APBS [3] where negatively charged regions are represented in red, neutral regions in white, and positively charged regions in blue (± 5 kT/e). Locations of alkaline surface residues are indicated. B) Ribbon diagram with highlighted residue side chains represented as sticks. Residues Asn87, Lys88, His90, and Trp91 of the highly conserved Asn-Lys-X-His-Trp motif, are shown in orange, residues Lys38, Trp39, Ala83, and Tyr84 are shown in green. Pspto_3016 is shown in the same orientation as panel A. C) Ribbon diagram of MotCF (PDB ID 1KAF; residues 105–211 displayed) shown in the saddle orientation with highlighted putative DNA binding residues Tyr134, Arg135, and Lys166 in yellow. D) Ribbon diagram of Pspto_3016 in the saddle orientation with residues Trp22, Lys24, and Lys65 highlighted in yellow. E) Pspto_3016 ConSurf [2] representation shown with spheres, using the aligned Pfam PF04237 sequences clustered from the CLANS [13] sub-family. Conserved surface residues Arg32, Lys38, Trp39, Lys65, Ala83, Tyr84, His85, Met86, Asn87, Lys88, and Trp91 are indicated. EF) ConSurf representation of YjbR from Escherichia coli using the corresponding CLANS sub-family sequences. YjbR was aligned to Pspto_3016 in the same orientation as panels D and E. Residues N74, K75, and H77 of the Asn-Lys-X-His-Trp motif are indicated on the surface and Trp78 is buried. Additional conserved residues His21, Arg61, and His72 are indicated.
