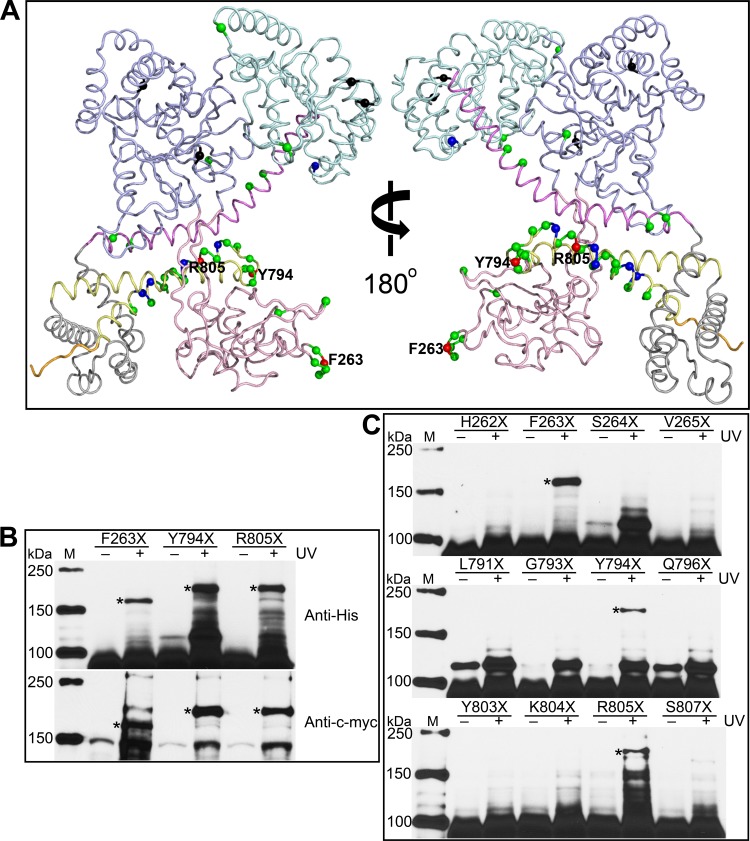
Fig 4.
Phe263, Tyr794, and Arg805 are found on the SecA dimer interface. (A) Residues investigated by in vivo photo-cross-linking are labeled on the ecSecA nuclear magnetic resonance (NMR) structure (PDB ID, 2VDA) with the signal peptide masked. The structure presentation is colored according to SecA domain organization as follows: NBD, light blue; IRA2, pale cyan; PBD, light pink; SD, violet; WD, gray; IRA1, pale yellow; and CTD, orange. The colored spheres show the positions of the amber mutations. Black spheres indicate the variants expressed as insoluble proteins, and green, blue, and red spheres indicate the variants yielding no, weak, and strong cross-linked SecA dimer, respectively. Residues showing strong cross-linking signals are labeled. (B) Cross-linked SecA F263X, Y794X, and R805X dimer bands were confirmed by Western blotting with anti-His and anti-c-myc antibodies. (C) No cross-linked dimer bands form when the flanking residues of Phe263, Tyr794, or Arg805 are replaced with pBpa and UV irradiated. Cross-linked dimer bands are indicated with asterisks. Lanes M, protein molecular mass markers, with molecular masses shown on the left.