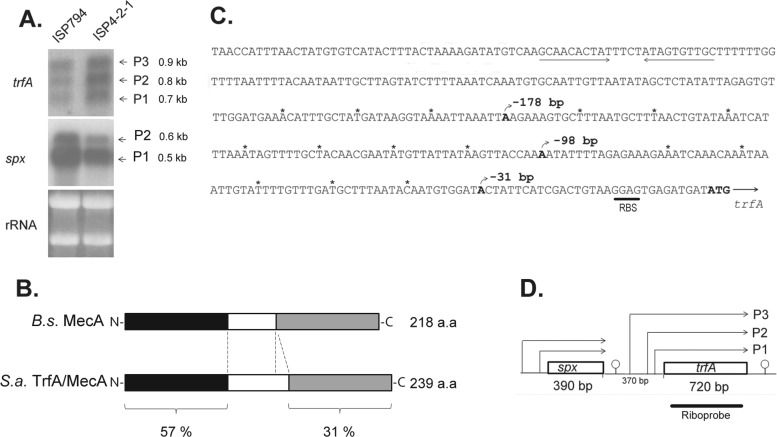
Fig 4.
Transcriptional analysis of trfA. (A) Northern blot analysis of trfA and spx in strains ISP794 and ISP4-2-1, using 32P-radiolabeled RNA trfA- and spx-specific probes. The arrows indicate the trfA (0.7-kb, 0.8-kb, and 0.9-kb) and spx (0.6-kb and 0.5-kb) transcripts. Ethidium bromide-stained rRNA from the agarose gel prior to blot transfer is shown as a loading control. (B) Schematic representation of B. subtilis (B.s.) MecA and S. aureus (S.a.) TrfA/MecA proteins. Throughout the text, we refer to S. aureus TrfA to avoid confusion with S. aureus mecA, a gene unrelated to the MecA adaptor protein of B. subtilis and encoding an alternative penicillin binding protein responsible for the MRSA phenotype in the organism. The N-terminal and C-terminal protein regions are depicted in black and gray and show 57% and 31% protein identity, respectively. The linker region (white) is smaller in B. subtilis MecA. (C) Sequence of the trfA promoter region. The three different nucleotides corresponding to transcriptional start sites detected by 5′RACE are shown in boldface, and the rho-independent transcriptional terminator of spx is underlined. RBS, ribosome binding site. Asterisks mark each 10-nucleotide region. (D) Schematic diagram showing spx and trfA gene transcription organization. The arrows indicate spx or trfA transcripts produced from the corresponding promoters. Predicted rho-independent transcriptional terminators are shown.