Figure 2. Differentially Methylated Regions (DMRs) during pollen development.
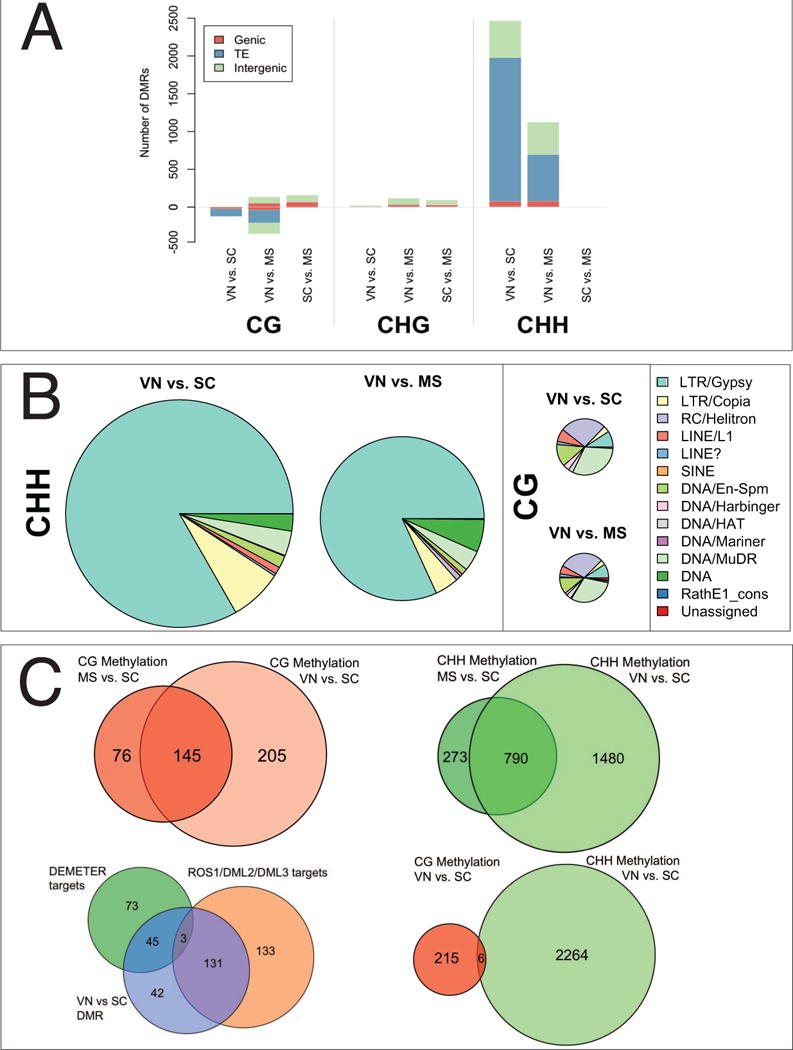
(A) DMRs were detected in a pairwise manner by comparing the bisulfite-sequence profiles from each of the three pollen cell types (vegetative nucleus-VN, sperm cell-SC, and microspore-MS) in each methylation context (CG, CHG, CHH). Annotated features (Genic, TE and Intergenic) overlapping one or more DMR in each cell type and methylation context were identified using TAIR10 annotation. Bars represent the number of DMRs overlapping each feature class. Where a DMR overlaps two or more features each feature is counted once. (B) Scaled distribution of transposon classes overlapping DMRs in the VN. TEs that matched each DMR were identified. Where a DMR overlaps two or more TE superfamilies each overlap is counted once. DMRs that lost CG methylation in the VN were enriched for class II DNA transposons, while DMRs that lost CHH methylation in sperm cells were enriched for class I LTR/gypsy transposons. There were very few CHG DMRs (data not shown) and these did not overlap transposons. (C) CG DMRs (red, upper left) and CHH DMRs (green, upper right) were similar in pairwise comparisons between the VN and the microspore, and the VN and the SC. CG DMRs in the VN (blue, bottom left) overlap with DMRs detected between WT endosperm and dme endosperm (green, bottom left), which are targets of DEMETER (Hsieh et al., 2009), and with DMRs between inflorescence and ros1/dml2/dml3 inflorescence (Lister et al., 2008) which are targets of ROS1 and its homologs (orange, bottom left). In the VN, CG DMRs (pink, bottom right) and CHH DMRs (green, bottom right) do not overlap.
