Figure 6. DNA methylation at hypervariable recurrent epialleles.
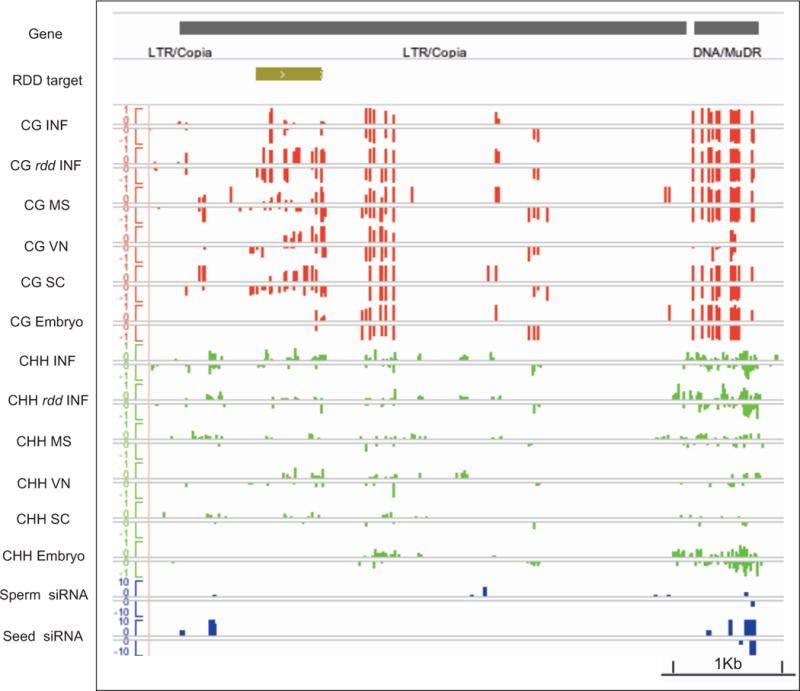
100 hypervariable epialleles gain DNA methylation recurrently in plants propagated by single seed descent (Becker et al., 2011; Schmitz et al., 2011). Many are targets of ROS1 and its homologs DML1 and DML2 (RDD). An example is shown (ATCOPIA51, At4g09455), along with a neighboring MuDR element for comparison. Tracks represent the RDD target region, and methylation levels in CG and CHH contexts in microspores (MS), vegetative nucleus (VN), and sperm cells (SC), along with inflorescence (INF) and embryo. CG methylation at the RDD target site is found in rdd triple mutant inflorescence (rdd INF) (Lister et al., 2008) and in pollen, but not in inflorescence or embryo. Small RNA from sperm cells (Slotkin et al., 2009) and seed (Lu et al., 2012) are also shown.
