Figure 7. Genome reprogramming during pollen development.
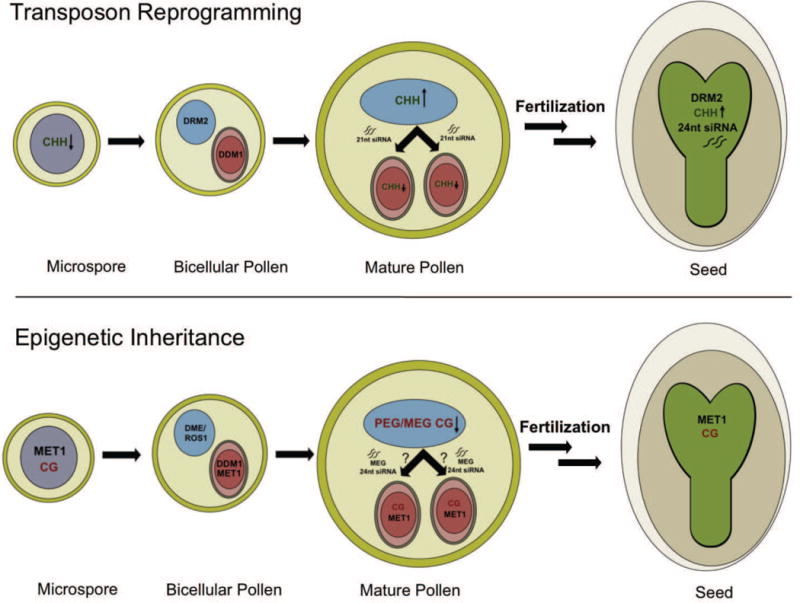
Differential expression of DRM2, MET1, ROS1, DME and DDM1 is depicted in bicellular pollen and persists in tricellular and mature pollen after the vegetative nucleus (VN, blue) and sperm cells (SC, red) differentiate (not shown). This results in reprogramming of transposons, imprinted genes and epialleles, as shown. Transposon reprogramming. DRM2 is down regulated in the microspore and sperm cells, so that CHH methylation is lost from retrotransposons, and is only restored after fertilization in the embryo (green), guided in part by maternal 24nt siRNA. DRM2 restores CHH methylation in the VN, guided by pollen 24nt siRNAs. In the vegetative cell, the chromatin remodeler DDM1 is lost, and retrotransposon activation generates 21nt siRNA that accumulate in sperm cells (arrow). Epigenetic inheritance. In the VN the DNA glycosylases DME and ROS1 target specific transposons for demethylation, including those that flank imprinted genes. In SC, CG methylation is maintained, and 24nt siRNA accumulate specifically from transposons that flank Maternally Expressed imprinted Genes (MEGs). These 24nt siRNAs may arise in the VN, resembling 21nt retrotransposon siRNA in this respect. A similar mechanism targets recurrent epialleles in pollen, contributing to their sporadic occurrence and to their subsequent inheritance in the embryo.
