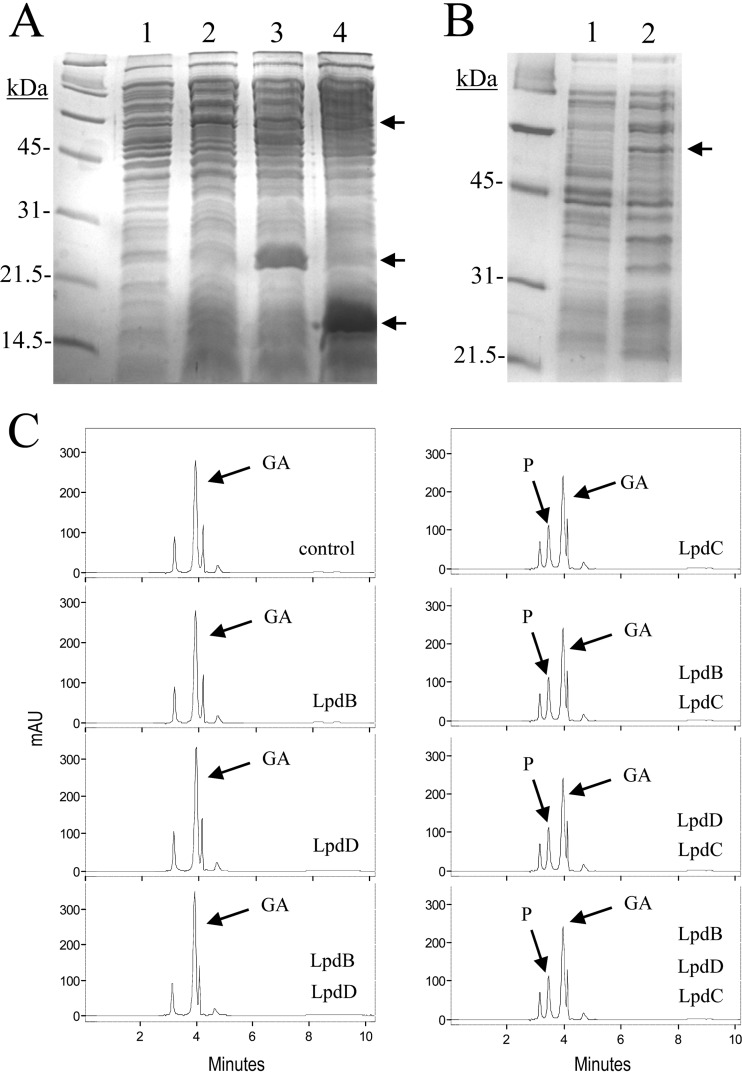
Fig 3.
Pyrogallol production by cell extracts from recombinant E. coli cells harboring lpdB, lpdC, and lpdD genes. (A and B) SDS-PAGE analysis of cell extracts of IPTG-induced cultures of E. coli DH5α bearing recombinant pIN-III-A3 plasmids for LpdB, LpdC, and LpdD protein production. (A) LpdC, LpdB, and LpdD production (15% gel). Lane 1, E. coli DH5α(pIN-III-A3); lane 2, E. coli DH5α(pIN-lpdC); lane 3, E. coli DH5α(pIN-lpdB); lane 4, E. coli DH5α(pIN-lpdD). (B) LpdC production (12% gel). Lane 1, E. coli DH5α(pIN-III-A3); lane 2, E. coli DH5α(pIN-lpdC). The arrows indicate the overproduced proteins. The gels were stained with Coomassie blue. Molecular mass markers are located on the left. (C) Gallate decarboxylase activity of E. coli DH5α cell extracts harboring lpdB, lpdC, and lpdD genes. HPLC chromatograms of E. coli cell extracts, at the same total protein concentration, incubated in 3 mM gallic acid for 1 h: pIN-III-A3 (control), pIN-III-A3 plus pIN-lpdB (LpdB), pIN-III-A3 plus pIN-lpdD (LpdD), pIN-III-A3 plus pIN-lpdB and pIN-lpdD (LpdB LpdD), pIN-III-A3 plus pIN-lpdC (LpdC), pIN-III-A3 plus pIN-lpdB and pIN-lpdC (LpdB LpdC), pIN-III-A3 plus pIN-lpdC and pIN-lpdD (LpdC LpdD), and pIN-lpdB plus pIN-lpdC and pIN-lpdD (LpdB LpdC LpdD). The gallic acid (GA) and pyrogallol (P) detected are indicated. Chromatograms were recorded at 280 nm. mAU, milli-absorbance units.