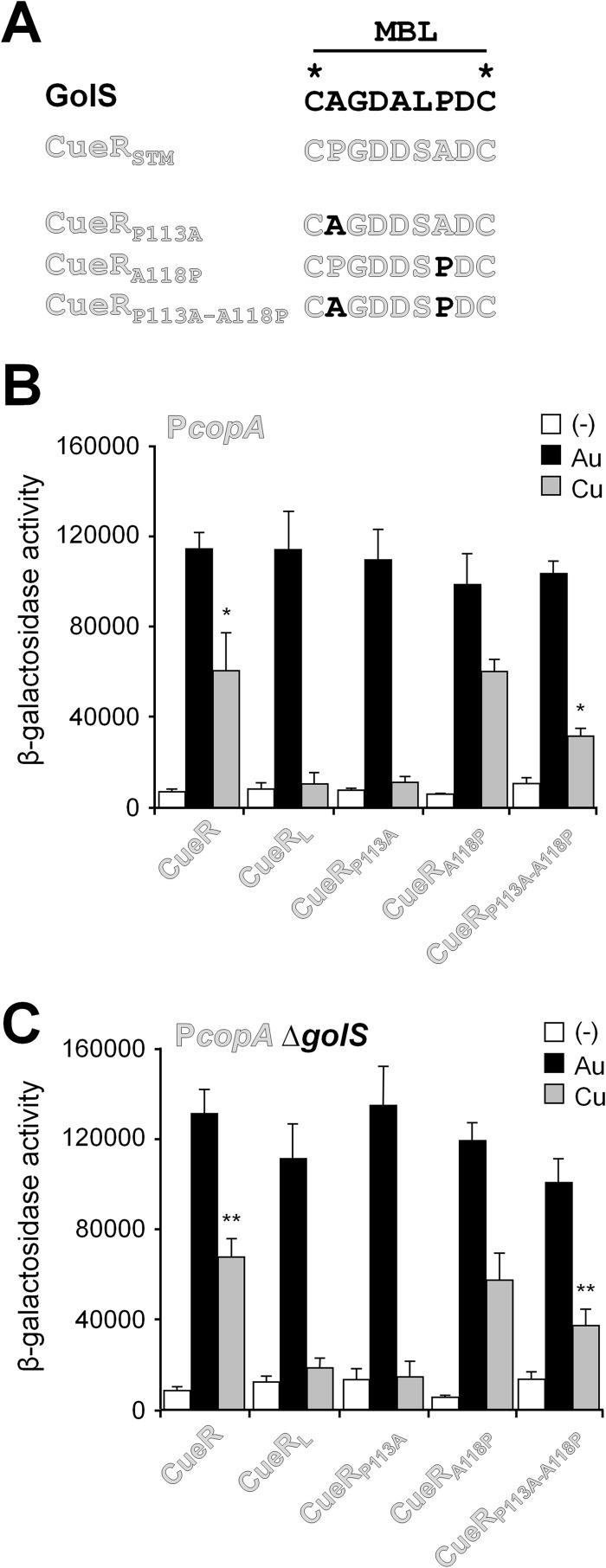
Fig 5.
Analysis of the roles of residues at positions 113 and 118 in Salmonella CueR. (A) Sequence of the MBL of GolS, CueR, and the mutant alleles CueRP113A, CueRA118P, and CueRP113A A118P. (B and C) β-Galactosidase activity from a copA::lacZ transcriptional fusion expressed from a reported plasmid by cells carrying cueR (CueR) or the mutant alleles coding for CueRL, CueRP113A, CueRA118P, or CueRP113A A118P in an otherwise wild-type (B) or ΔgolS (C) genetic background. Cells were grown overnight in LB broth without (−) or with the addition of 40 μM AuHCl4 (Au) or 1 mM CuSO4 (Cu). The data correspond to mean values of four independent experiments performed in duplicate. Error bars correspond to the standard deviations. Differences in β-galactosidase activity values from the wild-type CueR and CueRP113A A118P in the presence of Cu were statistically significant (*, P = 0.029; **, P = 0.001). No significant differences were observed from wild-type CueR and CueRA118P in the presence of Cu.