Fig 5.
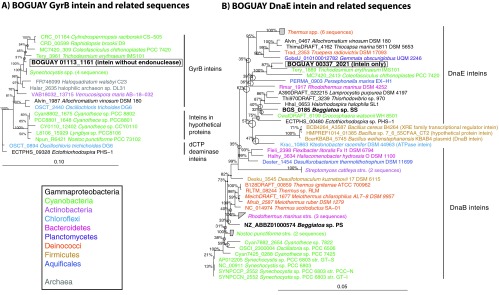
Inferred maximum-likelihood phylogeny of the putative BOGUAY GyrB and DnaE inteins. Relatives of the BOGUAY sequences (highlighted by boxes) were identified by BLASTX searches of the GenBank nr and JCVI IMG/ER databases. The first 100 hits were used for initial neighbor-joining trees, which were pruned to focus on the most closely Beggiatoa-related sequences before further analysis. Inferred amino acid sequences were aligned in MEGA5 (10) using MUSCLE (11). Maximum-likelihood phylogenies were inferred in ARB (13) with RAxML rapid bootstrapping (14), using the PROTMIX rate distribution model and WAG amino acid substitution model. Five hundred bootstraps were used for panel A, and 1,000 were used for panel B. Numbers at nodes represent bootstrap values for each node, and circles indicate uncertain branch points. The scale bar represents amino acid changes per position. Bayesian trees are presented for comparison in Fig. S6 in the supplemental material.
