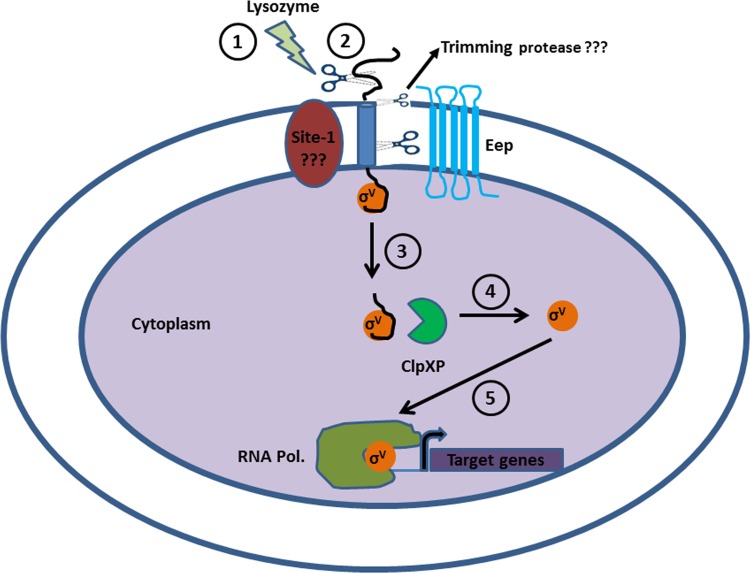
Fig 6.
A model for the regulated intramembrane proteolysis (RIP) of RsiV. A series of proteolytic events leads to the release and activation of SigV from its anti-sigma factor RsiV. (1) E. faecalis perceives a stress signal, which in this case is lysozyme. (2) This leads to the cleavage of RsiV by the putative site 1 protease. Based on data from the experiment whose results are shown in Figure 3 and from reference 28, E. faecalis and other Gram-positive bacteria, such as B. subtilis, possess an additional putative trimming protease activity that prepares the processed RsiV for further proteolytic targeting by Eep. (3) Eep degrades the site 1 protease-processed and -trimmed RsiV, leading to the release of SigV into the cytoplasm. (4) ClpXP cytoplasmic protease further degrades RsiV to release active SigV. (5) SigV initiates the binding of the RNA polymerase upstream from specific genes that confer lysozyme resistance.