LETTER
In this era of effective pneumococcal conjugate vaccines, simple and inexpensive methods are desirable for determining capsular serotype (st) distributions. Although a multiplexed conventional PCR (cmPCR) approach is generally reliable (1–4) (updates at www.cdc.gov/ncidod/biotech/strep/pcr.htm), the key serotypes 19A and 19F have presented problems (5, 6). The wzy19A- and wzy19F-encoded oligosaccharide repeat unit polymerases putatively provide the basis of the structural difference between the st19A and st19F capsules (7). Recently, we reported an st19F isolate recovered in Canada (strain 2584-0819F) that yielded a false-positive cmPCR type 19A (cmPCR-19A) result and a false-negative cmPCR-19F result (5). We redesigned our st19A primers in order to avoid further false-positive PCR-19As. Due to the similarity of the strain 2584-0819F wzy to the common st19A wzy, we were unable to design a new assay that would encompass strain 2584-0819F and the most commonly globally encountered 19F wzy gene represented by GenBank accession number CR931678 (unpublished data). More recently, a related circumstance described two cmPCR-19F isolates that expressed an st19A capsule (6).
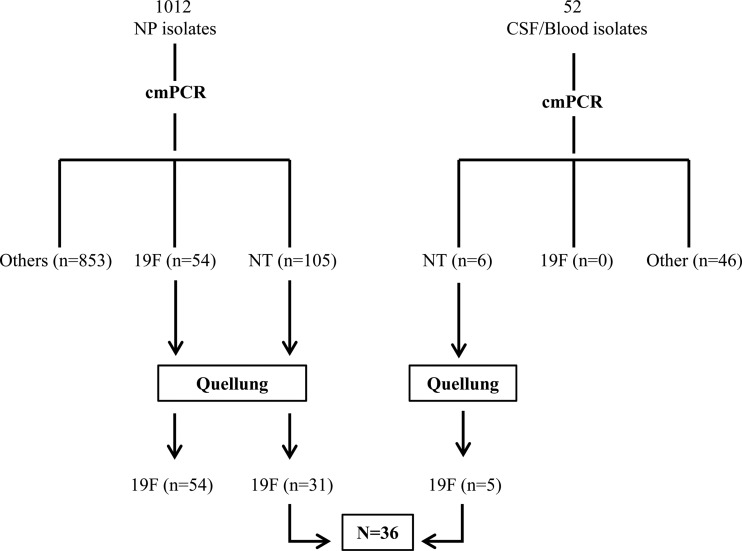
Here we describe a third variant wzy gene that was common within a collection of 1,064 pneumococcal isolates recovered in Brazil between 2010 and 2011 (cities of Goiânia and Salvador) (Fig. 1). Using our standard assay (4), the 1,012 carriage isolates included 54 cmPCR-19F isolates and 105 cmPCR-nontypeable isolates. All 54 cmPCR-19F isolates were st19F employing the Quellung reaction (Quellung-st19F). Of the 105 cmPCR-nontypeable carriage isolates, 31 were Quellung-st19F. The 52 invasive isolates included none that were cmPCR-19F; however, 5 of the 6 cmPCR-nontypeable isolates were Quellung-st19F. In total, 36 of the 90 Quellung-19F isolates were cmPCR nontypeable.
Fig 1.
Recovery of 36 serotype 19F isolates that were cmPCR nontypeable from carriage and invasive sources. Six hundred twenty pneumococcal carriage isolates were obtained from a survey conducted in Goiânia city from December 2010 through February 2011 (each isolate from a separate child). Of these isolates, 13 were cmPCR nontypeable (cmPCR-NT) and serotype 19F. Three hundred ninety-two isolates from 200 children that were less than 5 years of age were collected in Salvador city during 2008 (multiple swabs from some children at 3-month intervals), 20 of which were cmPCR-NT and serotype 19F (18 of which were from a separate child). All 52 isolates recovered from CSF or blood represented independent infections occurring in Salvador city between 2008 and 2010. All 36 cmPCR-NT, serotype 19F isolates, and, in addition, the previously described 19F sequence variant 2584-08 (5), were PCR typeable using a new primer set derived from their wzy sequences (GenBank accession numbers KC690152 and FJ829071). NP, nasopharyngeal; CSF, cerebrospinal fluid.
Sequencing the 1,335-bp wzy genes from 4 of the 35 cmPCR-nontypeable Quellung-st19F isolates (two invasive, two carriage) revealed them to be identical and to share 98.8% identity with the wzy structural gene from 2584-0819F (6). Thus, within Brazil, there appears to be a second major wzy allele (GenBank accession number KC690152) represented by 40% (36/90) of st19F isolates (Fig. 1). We designed new cmPCR primers to target this new wzy variant (primers 19FvarF [GACAATTCTGGTTGACTTGTTGATTTTG] and 19FvarR [CTACCAAATACCTCACCAGCTTCC]). All 36 serotype 19F isolates previously found to be cmPCR nontypeable, as well as the previously described 2584-0819F (6), were positive for a 585-bp amplicon following amplification with these primers. These primers do not target known 19A wzy genes.
In the recent study (6), 168 of 170 cmPCR-19F isolates found through using the standard CDC 19F PCR assay were st19F using the Quellung reaction, while the remaining two isolates were st19A. It is logical to assume that there will be a low error rate when employing cmPCR for deducing st19A, st19F, and possibly other serotypes. Until sequence signatures that dictate differences between the 19F and 19A capsular structures are deduced, better resolution of these two serotypes is problematic. Nonetheless, available data indicate that primer 19FvarF and 19FvarR-directed PCR may be useful for detecting a major st19F wzy variant within Brazil and elsewhere. There is a continued need for monitoring cmPCR-based serotype distribution data through using antibody-based serotyping as a quality control measure.
Nucleotide sequence accession number.
The newly determined wzy sequence has been deposited in GenBank under accession number KC690152.
ACKNOWLEDGMENTS
This study was partially supported by the Research Support Foundation for the State of Bahia (FAPESB; 1431040054051), the National Institutes of Health (D43 TW TW00919 and TW 007303), and a Fogarty International Center Global Infectious Diseases Research Training Program grant, National Institutes of Health, to the University of Pittsburgh.
We thank the laboratory staff at the CDC, especially Zhongya Li, for technical support.
Footnotes
Published ahead of print 8 May 2013
REFERENCES
- 1. de O Menezes AP, Campos LC, dos Santos MS, Azevedo J, dos Santos RC, Carvalho MDG, Beall B, Martin SW, Salgado K, Reis MG, Ko AI, Reis JN. 2011. Serotype distribution and antimicrobial resistance of Streptococcus pneumoniae prior to introduction of the 10-valent pneumococcal conjugate vaccine in Brazil, 2000-2007. Vaccine 29:1139–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dias CA, Teixeira LM, Carvalho MDG, Beall B. 2007. Sequential multiplex PCR for determining capsular serotypes of pneumococci recovered from Brazilian children. J. Med. Microbiol. 56:1185–1188 [DOI] [PubMed] [Google Scholar]
- 3. Morais L, Carvalho MDG, Roca A, Flannery B, Mandomando I, Soriano-Gabarro M, Sigauque B, Alonso P, Beall B. 2007. Sequential multiplex PCR for identifying pneumococcal capsular serotypes from South-Saharan African clinical isolates. J. Med. Microbiol. 56:1181–1184 [DOI] [PubMed] [Google Scholar]
- 4. Carvalho MDG, Pimenta FC, Jackson D, Roundtree A, Ahmad Y, Millar EV, O'Brien KL, Whitney CG, Cohen AL, Beall BW. 2010. Revisiting pneumococcal carriage by use of broth enrichment and PCR techniques for enhanced detection of carriage and serotype. 2010. J. Clin. Microbiol. 48:1611–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pimenta FC, Gertz RE, Jr, Roundtree A, Yu J, Nahm MH, McDonald RR, Carvalho MDG, Beall BW. 2009. Rarely occurring 19A-like cps locus from a serotype 19F pneumococcal isolate indicates continued need of serology-based quality control for PCR-based serotype determinations. J. Clin. Microbiol. 47:2353–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Siira L, Kaijalainen T, Lambertsen L, Nahm MH, Toropainen M, Virolainen A. 2012. From Quellung to multiplex PCR, and back when needed, in pneumococcal serotyping. J. Clin. Microbiol. 50:2727–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aanensen DM, Mavroidi A, Bentley SD, Reeves PR, Spratt BG. 2007. Predicted functions and linkage specificities of the products of the Streptococcus pneumoniae capsular biosynthetic loci. J. Bacteriol. 189:7856–7876 [DOI] [PMC free article] [PubMed] [Google Scholar]