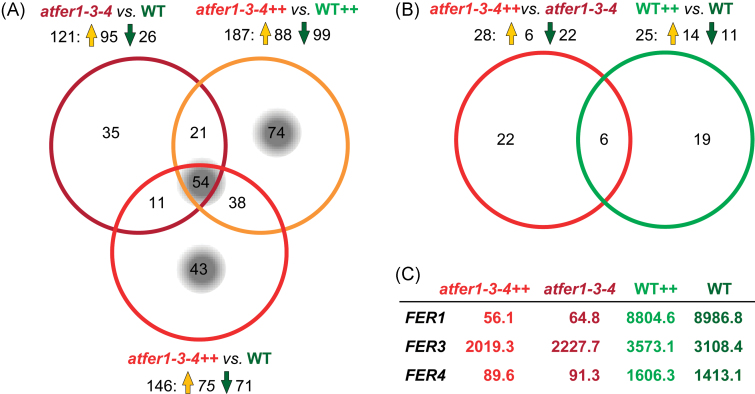
Fig. 1.
Changes in Arabidopsis flower transcripts of atfer1-3-4 mutants and the wild type. Overlapping regulation is shown by Venn diagrams. Numbers of regulated genes are given; up- and down-regulation is indicated by orange and green arrows, respectively (for fold changes see Supplementary Table S5 at JXB online). (A) Differentially expressed genes (P-value ≤0.1) in flowers of atfer1-3-4 mutants compared with the Col-0 wild type (WT) under Fe-sufficient and Fe excess (++) conditions. In total, 276 genes were significantly regulated when comparing atfer1-3-4 versus WT, atfer1-3-4++ versus WT++, and atfer1-3-4++ versus WT. Gene groups of interest discussed in the text are highlighted by a grey background. (B) Transcript changes (P-value ≤0.2) induced by Fe excess (++) in atfer1-3-4 and the wild type using the respective Fe-sufficient conditions as control. Note that all genes regulated in atfer1-3-4++ versus atfer1-3-4 and WT++ versus WT are also differentially expressed in one of the comparisons depicted in (A). (C) Mean signal values (arbitrary units) of mRNA from mutated ferritin genes (AtFER1, AtFER3, and AtFER4) in flowers of all samples analysed.