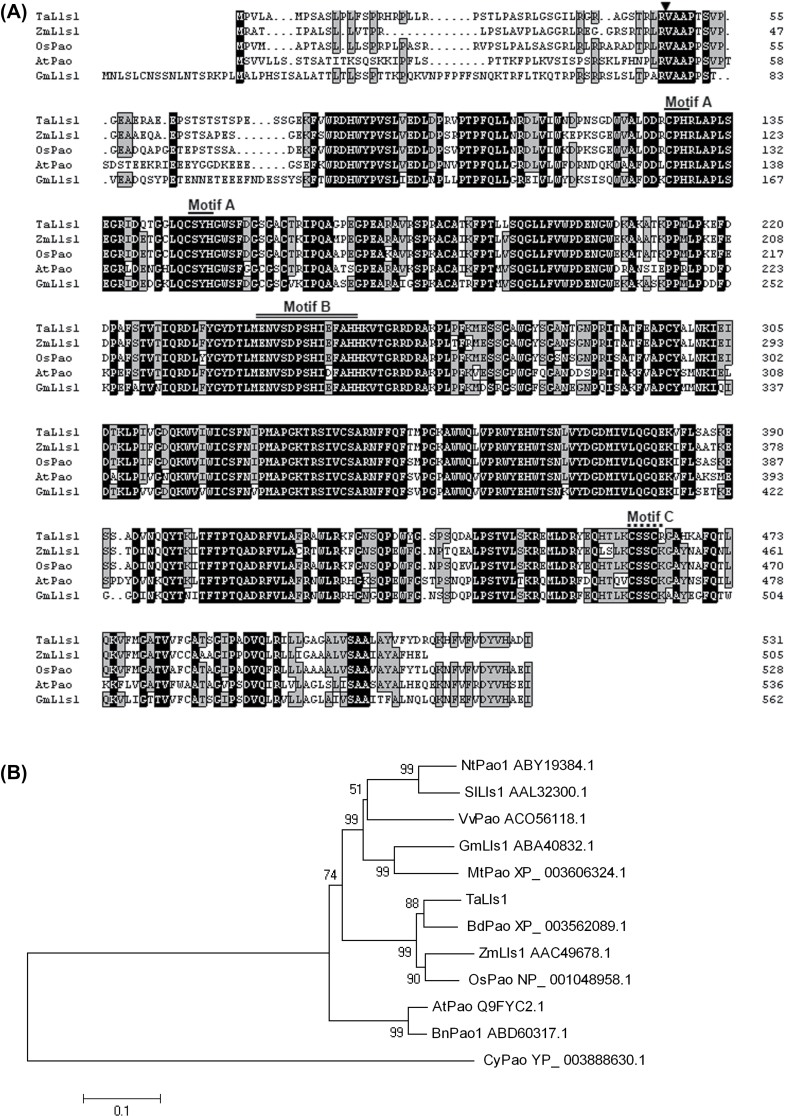
Fig. 1.
Multi-sequence alignment and phylogenetic analysis of TaLls1 and other members of the PaO family. (A) Multiple alignment of amino acids. Identical and similar amino acid residues are shaded in black and light grey, respectively. The arrow indicates the cleavage site of the chloroplast peptide; Motif A, indicated by a single line, is the Rieske iron-binding motif; Motif B, indicated by a double line, is the mononuclear iron-binding site; Motif C, indicated by a dashed line, is the conserved CxxC sequence. (B) Phylogenetic analysis of TaLls1 and other PaO family members using MEGA 4.1 software. Branches are labelled with the protein names and GenBank accession numbers. Ta, Triticum aestivum L; Os, Oryza sativa; Zm, Zea mays; At, Arabidopsis thaliana; Nt, Nicotiana benthamiana; Sl, Solanum lycopersicum; Gm, Glycine max; Bn, Brassica napus; Mt, Medicago truncatula; Vv, Vitis vinifera; Cy, Cyanothece sp.