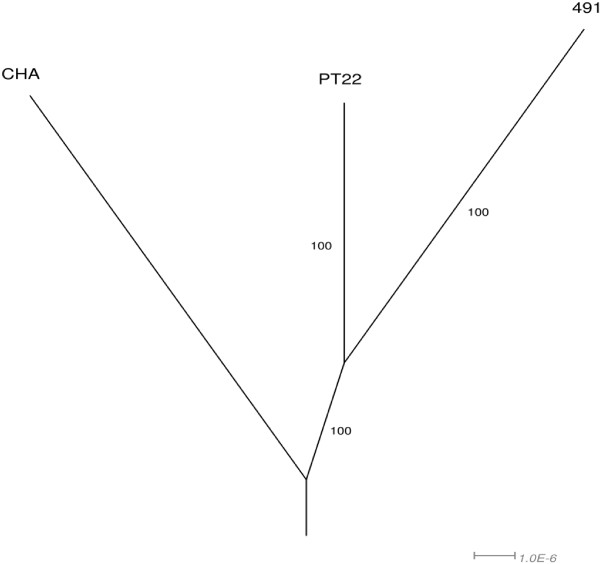
Figure 3.
Phylogenetic network for clone CHA isolates based on identified SNPs. All SNPs mentioned in Figure 1 were incorporated into three pseudosequences derived from the PAO1 reference sequence by the script SequenceReplacer (available on request from the authors). The network was produced using the uncorrected P distance measure with normalisation followed by the NeighbourNet algorithm in the program Splitstree [62]. The scale indicates the number of substitutions per site. Numbers on the branches are 100 bootstrap resampling values which give a measure of the confidence of the displayed tree topology. A network for clone TB is not shown as the isolates display up to two orders of magnitude less divergence than clone CHA strains, which cannot be visualised appropriately.