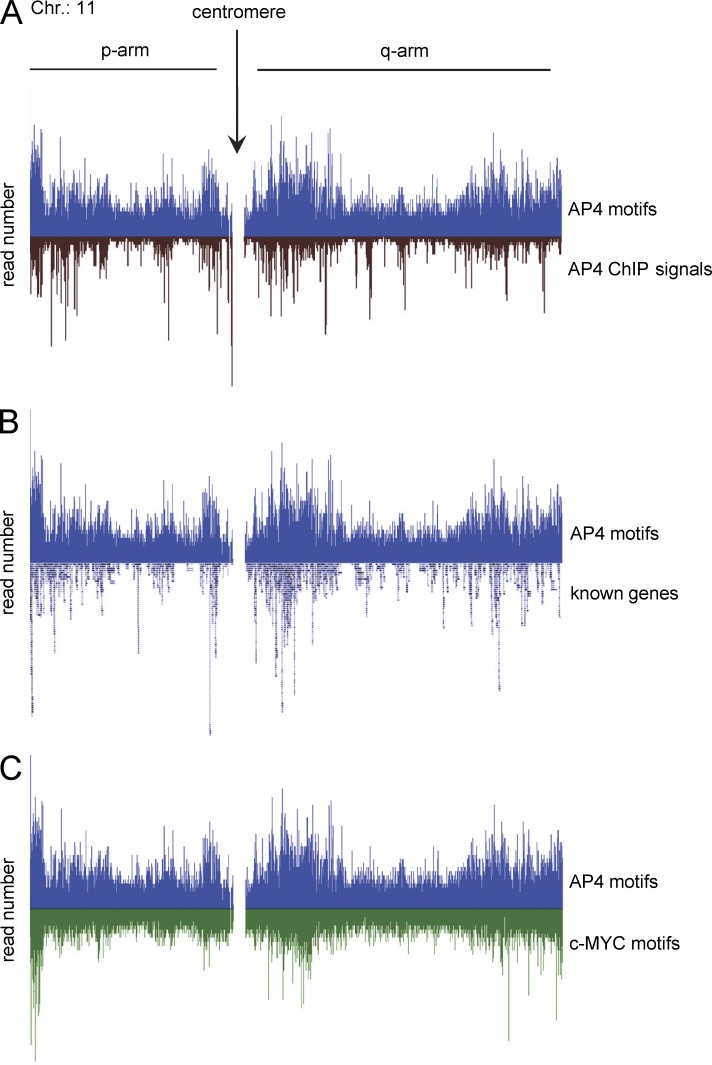
Figure 3.
Comparison of the distribution of AP4-binding motifs with ChIP signals, transcription units, and c-MYC motifs on chromosome 11. Gene distributions are shown according to UCSC annotation. Raw ChIP-seq reads are shown before peak calling. The transcription factor–binding motifs CAGCTG (AP4) and CACGTG (c-MYC) were mapped to the human genome (hg19). (A) AP4 motif distribution compared with AP4-derived ChIP signals. (B) Comparison of AP4-binding motif distribution with the location of known genes. (C) Comparison of the distribution of AP4- with c-MYC–binding motifs.