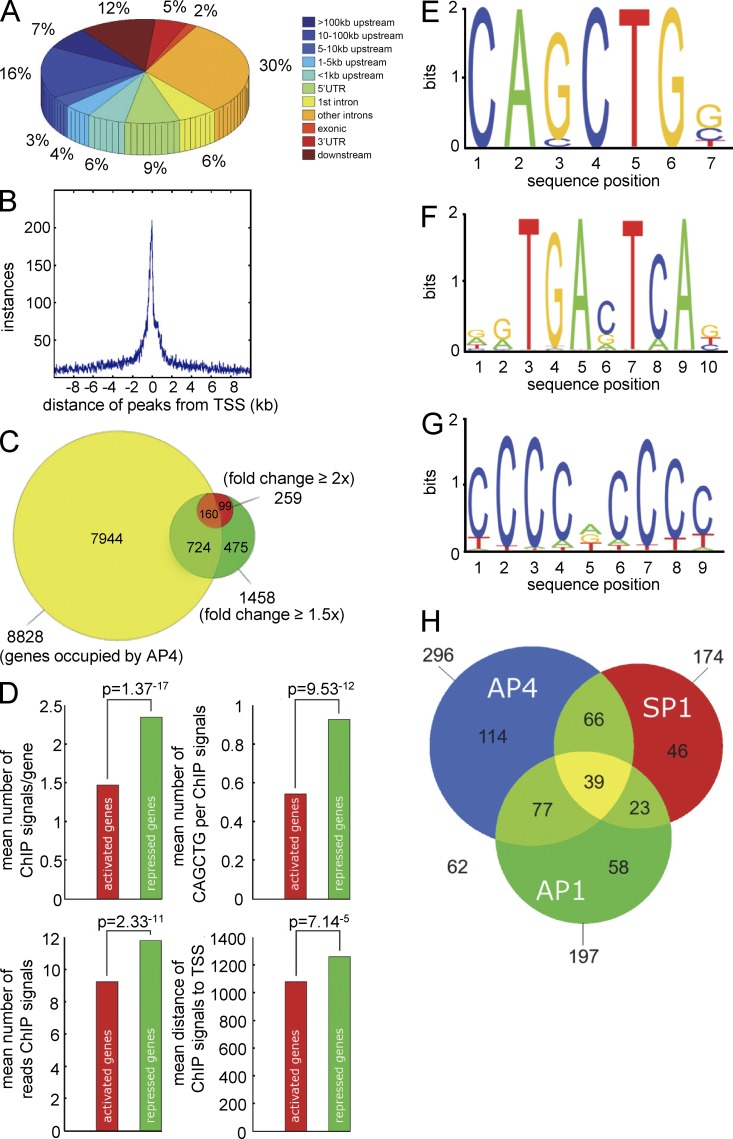
Figure 4.
Genome-wide analyses of AP4 occupancy. DLD-1/pRTR-AP4-VSV cells were treated with DOX for 24 h. After ChIP with an AP4-specific antibody, the coprecipitated DNA was subjected to next generation sequencing. For details of the ChIP-seq analysis, see Materials and methods. (A) Localization of called ChIP signals. ChIP signals were assigned using the closest TSS and mapped to specific genomic regions using UCSC annotations. (B) AP4-binding pattern in the vicinity of the TSS. ChIP signals located within a range of 10 kbp up- or downstream of the TSS of 10,636 genes were analyzed. (C) Proportion of differentially expressed genes with AP4 binding in the promoter region. Results of mRNA expression analysis (green and red) and AP4 occupancy were compared (yellow). (D) Comparison of AP4-derived ChIP signals at the promoters of AP4-induced and -repressed genes. Peaks located within 5 kbp up- and 3 kbp downstream of a TSS were considered. The p-values were calculated using a two-sample Kolmogorov-Smirnov test. (E) Refinement of the AP4-binding motif by MEME analysis of AP4 ChIP-seq results. 485 ChIP signals in close proximity to a TSS with at least 25 overlapping reads were included. (F and G) Specific enrichment of AP1 and SP1 sites in the vicinity of AP4 sites was determined by the MEME and Gibbs motif samplers. (H) Distribution of co-occurring motifs in proximity of 485 ChIP-seq peaks with at least 25 overlapping, AP4-derived reads. 62 ChIP signals did not contain any of the three motifs.