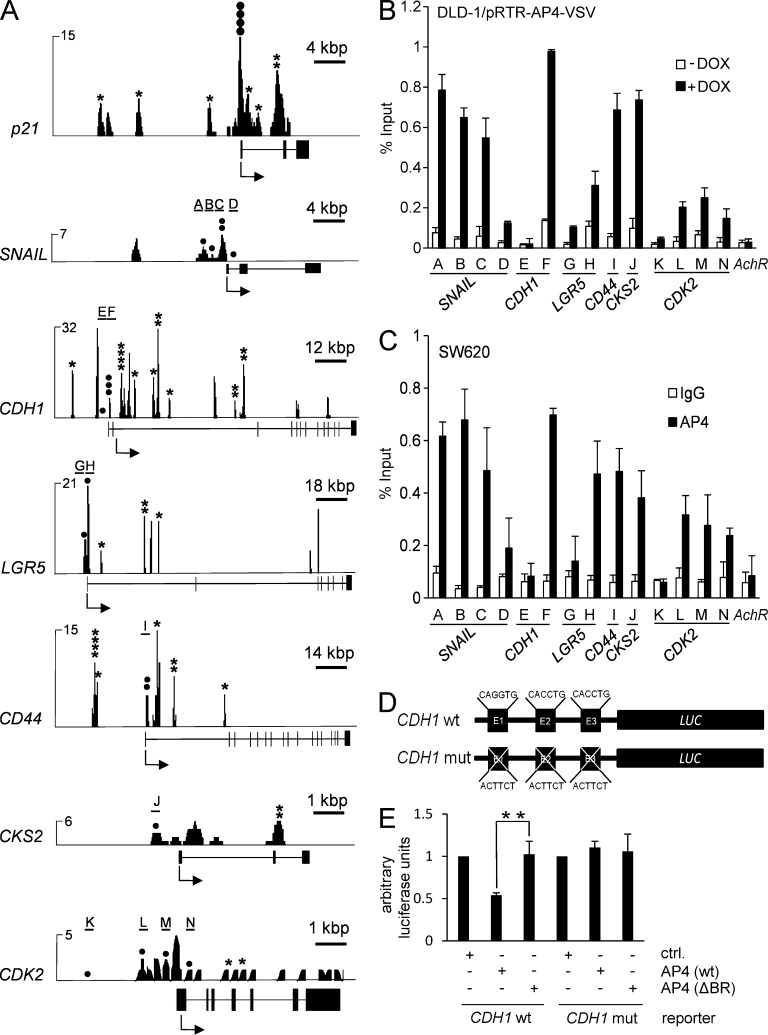
Figure 5.
Analysis of AP4 occupancy at selected promoters. (A) Representative ChIP-seq results for seven selected promoters. All results besides those for CDK2 are shown after peak calling. Asterisks represent bioinformatically predicted CAGCTG motifs. Closed circles represent CAGCTG motifs (CACCTG in the CDH1 promoter) analyzed by qChIP. Letters and horizontal bars represent the qPCR amplicons used for qChIP analysis in B. (B) DLD-1/pRTR-AP4-VSV cells were treated with DOX for 24 h and subjected to qChIP analysis with an AP4-specific or, as a reference, IgG antibody. (C) SW620 cells were subjected to qChIP analysis with an AP4-specific or, as a reference, IgG antibody. In B and C, the acetylcholine receptor (AchR) promoter, which lacks AP4-binding motifs, served as a negative control, and results are depicted as mean ± SD (n = 3). (D and E) Scheme of CDH1/E-cadherin reporters (wt = wild-type; mut = three CACCTG motifs mutated) that were used for the dual-luciferase assay (E) 48 h after transfection of the indicated reporters and the indicated AP4 constructs (ΔBR = deleted DNA binding region) into DLD-1 cells. Experiments in E were performed in triplicates (n = 3); error bars indicate SD; significance level as indicated: **, P < 0.01.