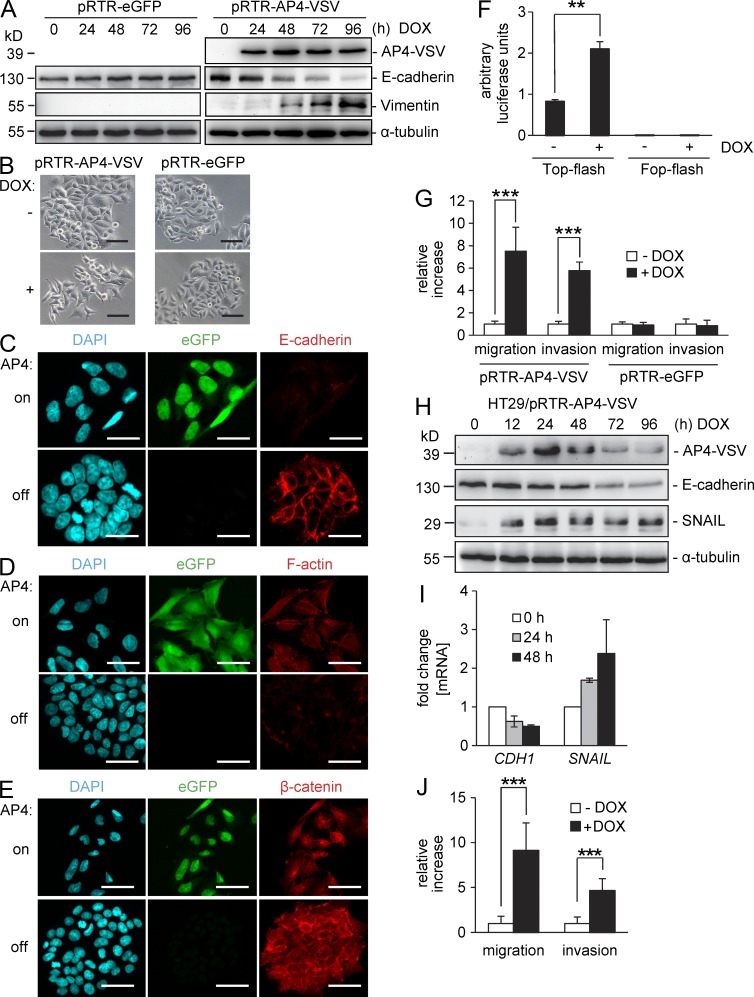
Figure 6.
Ectopic AP4 induces EMT in CRC cells. (A) A conditional AP4 allele was induced in DLD-1 cells by addition of DOX for the indicated periods. As a control, DLD-1 cells harboring a pRTR vector were treated with DOX. Protein lysates were subjected to immunoblot analysis for detection of the indicated proteins. (B) Representative phase-contrast pictures of DLD-1 cells 96 h after activation of a conditional AP4 allele or, as a control, of eGFP by addition of DOX. (C–E) Confocal microscopy of DLD-1 cells ectopically expressing AP4 after induction by DOX or the respective untreated control: E-cadherin and β-catenin were detected by indirect immunofluorescence. F-actin was visualized with Alexa Fluor 647–labeled Phalloidin. Cellular DNA was stained with DAPI. (B–E) Bars, 50 µm. (F) Dual-luciferase assay 24 h after transfection of the indicated TCF4 reporter constructs and simultaneous induction of conditional AP4 expression by addition of DOX. (G) Boyden chamber transwell assay of cellular migration or invasion in DLD-1 cells harboring a conditional AP4 allele or, as a control, the control vector only expressing eGFP. Cells were cultivated in the presence or absence of DOX for 96 h with serum reduction to 0.1% for the last 24 h. To analyze invasion, membranes were coated with Matrigel. After 48 h, cells were fixed and stained with DAPI. The mean number of cells in five fields per membrane was counted in three different inserts. Relative invasion or migration is expressed as the value of test cells to control cells. (H) A conditional AP4 allele was induced in HT29 cells by addition of DOX for the indicated periods. Protein lysates were subjected to immunoblot analysis for detection of the indicated proteins. (A and H) α-Tubulin detection served as a loading control. (I) qPCR analysis of RNA obtained at the indicated time points after AP4 activation. β-Actin was used for normalization. Results represent the mean ± SD (n = 3). (J) Boyden chamber transwell assay of cellular migration or invasion as in G in HT29 cells harboring a conditional AP4 allele. Experiments in F, G, and J were performed in triplicates (n = 3). Error bars indicate SD; significance level as indicated: **, P < 0.01; ***, P < 0.001.