Fig. 4.—
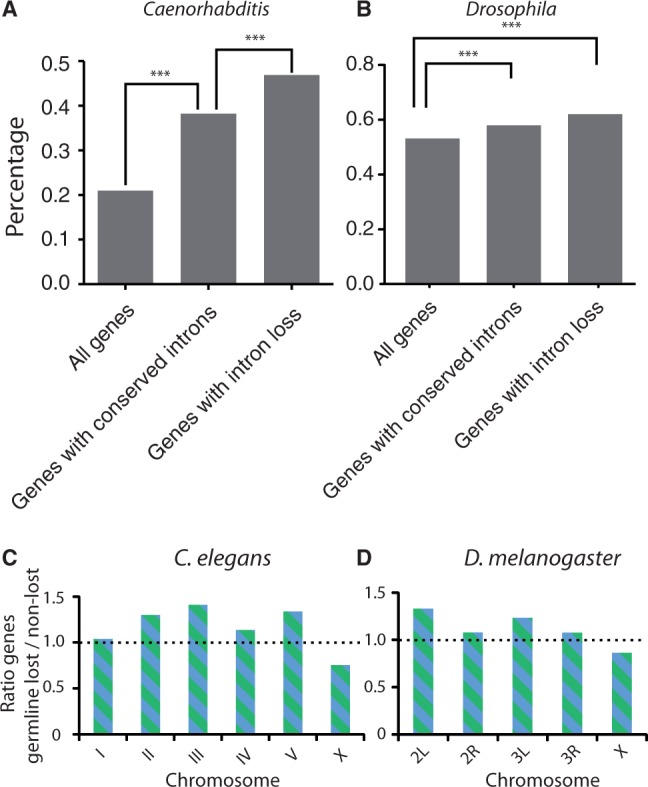
Increased likelihood of intron loss in germline-expressed genes in (A) Caenorhabditis elegans and (B) Drosophila melanogaster. Our criteria for conserved introns, selecting on highly conserved surrounding exons, enriches for germline-expressed genes (P < 0.001, χ2 test). Germline expression was highly overrepresented in the class of genes with associated intron loss (P < 0.001, χ2 test). ***P < 0.001. (C) Distribution of germline-expressed genes across the autosomes and the X-chromosome in C. elegans. For each chromosome, the ratio between germline-expressing genes that have lost at least one intron and genes that contain only retained introns is plotted, [(D as in C)] but now for D. melanogaster. We find the same outcome as for C. elegans: introns located in germline-expressing genes on X are less prone to be lost compared with introns located on the autosomes.
