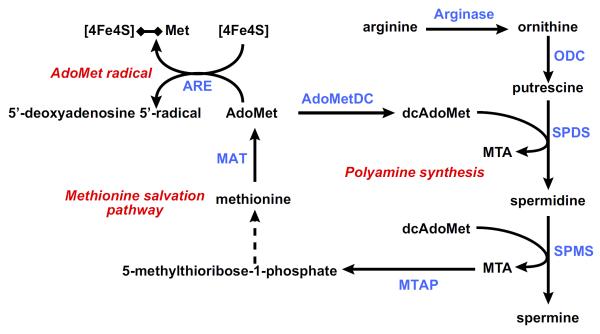
FIGURE 4.
Methionine metabolism: polyamine synthesis, methionine salvation pathway, and AdoMet radical reactions. To synthesize polyamines, AdoMet needs first to be decarboxylated, a reaction catalyzed by the enzyme AdoMet decarboxylase (AdoMetDC), to form decarboxylated AdoMet (dcAdoMet). The predominant polyamines in mammalian cells are spermidine (SPD) and spermine (SPM). These polyamines are made by sequential addition of aminopropyl groups from dcAdoMet, yielding 5′-methylthioadenosine (MTA) as a byproduct. SPD synthase (SPDS) catalyzes the transfer of the first aminopropyl group from dcAdoMet to putrescine to form SPD and MTA, whereas SPM synthase (SPMS) catalyzes the transfer of the second aminopropyl group to SPD to form SPM and a second molecule of MTA. MTA is metabolized through the methionine salvation pathway to regenerate AdoMet. The first reaction in this pathway is the cleavage of MTA by the enzyme MTA phosphorylase (MTAP) yielding adenine and 5-methylthioribose-1-phosphate, which is further metabolized to methionine and adenine to AMP. AdoMet may be also converted to 5′-deoxyadenosyl 5′-radical by a large family of AdoMet radical enzymes (ARE) and initiate a variety of radical chemical reactions. These enzymes share a CX3CX2C motif forming a characteristic [4Fe-4S] cluster. This [4Fe-4S] cluster binds AdoMet catalyzing its reductive cleavage to generate [4Fe-4S]-methionine and a 5′-deoxyadenosyl 5′-radical. ODC, ornithine decarboxylase; MAT, methionine adenosyltransferase.