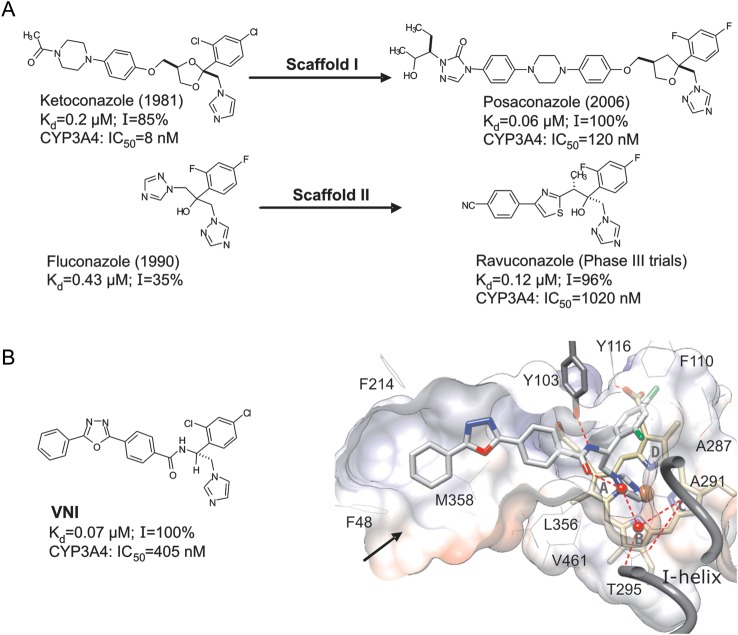
Figure 1.
Sterol 14α-demethylase (CYP51) inhibitory scaffolds of the antifungal drugs posaconazole and ravuconazole (derivatives of ketoconazole and fluconazole, respectively; A), and VNI (B). The numbers represent the comparative binding and inhibition parameters for Trypanosoma cruzi CYP51. Data denote the apparent dissociation constant (Kd), inhibition of the enzyme activity in vitro at equimolar ratio inhibitor/P450 (1-hour reaction; I), and median inhibitory concentrations (IC50) for inhibition of human CYP3A4, the cytochrome P450 that metabolized >90% of drugs in the human body [45]. B, VNI bound in the CYP51 active site [3gw9]. Shown is a slice through the surface of the binding cavity. VNI, the heme, and Y103 are depicted as stick models; other residues located within 4 Å are shown as lines. Water molecules are presented as red spheres. The H-bonds are displayed as red dashed lines. The substrate access channel entrance is marked with an arrow. Key features of VNI include an imidazole ring that directly coordinates to the enzyme heme iron (N3), a 2,4-dichlorinated β-phenyl ring that is projected into the deepest portion of the CYP51-substrate binding cavity, and a 3-ring linear polycycle positioned along the substrate access channel and connected to the rest of the inhibitor molecule through a carboxamide fragment with a chiral (+) amine carbon (R). The carbonyl oxygen of this fragment forms a hydrogen bond with the catalytic water molecule, thus connecting the inhibitor with the enzyme I-helix and, presumably, disrupting the CYP51 proton delivery route. The amide nitrogen repositions the side chain of Y103, which, in the enzymatically active protein, is H-bonded with the heme propionate. In total, VNI-CYP51 interactions include iron coordination, van der Waals contacts with 16 amino acid residues, and a H-bond network that disrupts 2 functionally essential protein regions.