Abstract
AIM: To investigate mitochondrial ATP 6 and 8 polymorphisms in the colon and ileum of patients with irritable bowel syndrome with diarrhea (IBS-D).
METHODS: Twenty-eight patients fulfilling the Rome III criteria for IBS-D and 28 healthy subjects were investigated. All study participants underwent screening colonoscopy and mucosal biopsies were obtained from the colon and/or terminal ileum. Genomic DNA was extracted from specimens based on standard protocols. Mitochondrial ATP (MT-ATP) 6 and 8 genes in specimens were polymerase chain reaction amplified and sequenced. Sequencing data were analyzed via Variant Reporter™ Software and compared with the reference sequence from Genbank (accession No. NC_012920) to indicate possible polymorphisms. The protocol was registered at www.clinicaltrials.gov as NCT01028898.
RESULTS: Twenty-five polymorphic sites of MT-ATP 6 and 8 genes were detected and 12 of them were missense mutations. A median of two polymorphic sites in MT-ATP genes was found in colon specimens of controls while a median of three polymorphic sites was noted in patients with IBS-D (Mann-Whitney test, P = 0.012). The variants of the colon and ileum specimens from the same subjects were identical in all but one case. Symptom duration in IBS was not found to be a significant factor associated with the mtDNA polymorphism (Spearman correlation, P = 0.592). The mitochondrial DNA change at 8860 was present in all cases of both groups. The frequency of the 8701 polymorphism was found to be the second most frequent; however, no statistical difference was noted between the groups (χ2 test, P = 0.584).
CONCLUSION: Patients with IBS-D have a higher incidence of MT-ATP 6 and 8 polymorphisms than healthy subjects, implying that the mtDNA polymorphism may play a role in IBS-D.
Keywords: Irritable bowel syndrome, Diarrhea, Mitochondrial ATP 6 gene, Mitochondrial ATP 8 gene, Polymorphism
Core tip: Mitochondrial DNA, the only source of extranuclear genome, was assessed by Camilleri’s group in a study of patients with functional gastrointestinal disorders in 2009 which was the first study exploring the possible role of mitochondrial DNA in irritable bowel syndrome (IBS). Up till now, few research efforts have focused on this topic and there is little knowledge about it. Present study revealed that mitochondrial ATP 6 and 8 genes were more frequently mutated in IBS with diarrhea than in healthy individuals. We believe this preliminary finding could help promote future research on mitochondrial DNA in IBS.
INTRODUCTION
Irritable bowel syndrome (IBS) is one of the most prevalent functional gastrointestinal disorders and affects 10%-20% of the population[1]. To date, the pathogenesis of IBS remains unclear.
Family[2,3] and twin[4] studies suggested that genetic changes may predispose individuals to IBS, although reported data are controversial[5]. Furthermore, some candidate nuclear genes are associated with IBS (serotonin transporter and receptor genes and inflammatory markers)[6]. Recently, mitochondrial DNA (mtDNA), the only source of extranuclear genes, was assessed by Camilleri et al[7] in a study of patients with functional gastrointestinal disorders. This initial study on the role of haplogroup H, 16519C>T and 3010G>A (located in the D-loop and the 16S rRNA gene) of mitochondrial DNA (mtDNA) indicated that the mitochondrial haplogroup and variations may be associated with gastrointestinal motor and sensory function in gastrointestinal disorders such as IBS. The human mitochondrial genome encodes 13 genes[8] for ATP subunits 6 and 8, and other polypeptides of respiratory complexes crucial for ATP production in mitochondria. ATP synthesis requires ATP synthase, of which subunits 6 and 8 are mtDNA encoded. Based upon this knowledge, we hypothesized that mitochondrial ATP (MT-ATP) 6 and 8 may be involved in the pathogenesis of IBS. In this study, we aimed to investigate the mitochondrial ATP (MT-ATP) 6 and 8 polymorphisms in the colon and ileum of IBS with diarrhea (IBS-D).
MATERIALS AND METHODS
Subjects
Twenty-eigh patients who satisfied Rome III criteria[1] for IBS-D and 28 healthy controls were enrolled at the Chinese PLA General Hospital between June 2009 and May 2011. Inclusion criteria for IBS with diarrhea were: (1) male and female individuals aged 18-60 years; (2) recurrent abdominal pain or discomfort associated with a change in stool frequency or form over a 3-mo period; (3) loose or watery stool more than 25% of the time, and hard stool less than 25% of the time; (4) symptom onset at least 6 mo prior to diagnosis; and (5) pain or discomfort at least 2 d a week during the screening period. Controls were healthy subjects undergoing routine health evaluation, with normal bowel movements (i.e., fewer than 3/d and more than 3/wk) and no gastrointestinal complaints in the previous year. Patients were excluded in both IBS-D and control groups if they: (1) were pregnant or lactating; (2) were found to have abnormal colonoscopic findings; (3) had symptoms of a significant clinical illness; (4) had previous gastrointestinal surgery; and (5) had diabetes or other diseases affecting gastrointestinal function. This study was approved by the Ethics Committee of Chinese PLA General Hospital and written informed consent was obtained from each subject. The protocol was registered at www.clinicaltrials.gov as NCT01028898. This study was conducted in compliance with the principles of the Declaration of Helsinki.
Endoscopy procedure
All study participants underwent screening colonoscopy with a CF-H260AI or CF-Q260AI colonoscope (Olympus, Tokyo, Japan); terminal ileum intubation was attempted when applicable. During withdrawal of the scope, the colonic mucosa was carefully visualized and biopsies of the sigmoid colon were obtained (FB-55U-1 biopsy forceps; Olympus, Tokyo, Japan). Terminal ileal biopsies were obtained from 10 patients with IBS-D.
All colonoscopies were performed at the Chinese PLA General Hospital and samples were stored in covered storage tubes at -20 °C before analysis.
Polymerase chain reaction and mtDNA sequencing
The MT-ATP 6 and 8 genes were polymerase chain reaction amplified and sequenced according to the mitoSEQr™ protocol (Applied Biosystems, Foster City, CA, United States).
Polymerase chain reaction reaction: Genomic DNA was extracted from specimens based on standard protocols[9]. Thermocycling conditions with the AB 9700 (Applied Biosystems, Foster City, CA, United States) were as follows: Heat activation at 96 °C for 5 min, followed by 40 cycles at 94 °C for 30 s, 60 °C for 45 s, 72 °C for 45 s; final extension at 72 °C for 10 min. Polymerase chain reaction (PCR) reaction clean-up was performed by adding 2 μL of ExoSAP-IT® (USB Corporation), incubating at 37 °C for 30 min and heat inactivation at 80 °C for 15 min.
Sequencing reaction and electrophoresis: A forward and reverse sequencing reaction mix was prepared from ready-to-use resequencing sets (Applied Biosystems, Foster City, CA, United States). The sequencing master mixes contained the M13 forward and reverse primers. Cycling conditions were: heat activation at 96 °C for 1 min, followed by 25 cycles at 96 °C for 10 s, 50 °C for 5 s, and 60 °C for 4 min. Sequencing reaction clean-up was performed. PCR products were electrophoresed using the AB 3730 DNA Analyzer (Applied Biosystems, Foster City, CA, United States).
Analysis of the sequences of MT-ATP 6 and 8 genes: Sequencing data were analyzed via Variant Reporter™ Software version 1.0 (Applied Biosystems, Foster City, CA, United States) and compared with the reference sequence from Genbank (accession No. NC_012920) to indicate possible polymorphisms. The corresponding changes of amino acid with missense polymorphisms were excerpted from MITOMAP (www.mitomap.org). For polymorphism not recorded in MITOMAP, HmtDB (http:// www.hmtdb.uniba.it: 8080/ hmdb/) was applied.
Statistical analysis
The χ2 and Student’s t test were used to analyze demographic data between control and IBS-D patients, wherever applicable. Since MT-ATP 6 and 8 genes overlap in the mtDNA genome, these two genes were considered as a single gene during analysis. The non-parametric Mann-Whitney U test was performed to analyze polymorphism frequency between groups. The Spearman rank correlation coefficient was used to determine the association between the presence of the mtDNA polymorphism and IBS-D duration. A P value of less than 0.05 was considered statistically significant. Statistical analysis was performed using SPSS software (Chicago, IL, United States).
RESULTS
Characteristics of study participants
The characteristics of study participants are summarized in Table 1. No significant difference was found in age or gender between patients with IBS-D and controls.
Table 1.
Demographic data and clinical profile of the participants
| Control (n = 28) | IBS (n = 28) | P value | |
| Age (yr) | 43.6 ± 1.9 | 41.6 ± 2.2 | > 0.051 |
| Gender (male:female) | 22:6 | 18:10 | > 0.052 |
| Disease duration (yr) | N/A | 3.6 ± 0.6 | N/A |
P values were obtained using t test (t = -0.682, df = 54, P = 0.498);
Pearson's χ2 test (χ2 = 1.400, df = 1, P = 0.237) between the control and patients with irritable bowel syndrome (IBS). Values expressed as mean ± SE. N/A: Not available.
mtDNA polymorphisms in colon and terminal ileum
The nuclear positions and base changes of the mtDNA polymorphism, as compared with the Genbank mtDNA sequence (accession No. NC_012920) are listed in Table 2. Twenty-five polymorphic sites in MT-ATP 6 and 8 genes were observed, of which 48% (12/25) were missense. No deletions or insertions were detected.
Table 2.
Summary of mitochondrial DNA mutations in the ATP 6 and 8 genes
| Nucleotide position | Base change (N>T) | mtDNA region | Amino acid change |
| 8392 | G>A | ATP8 | syn |
| 8394 | C>T | ATP8 | Missense |
| 8414 | C>T | ATP8 | Missense |
| 8459 | A>G | ATP8 | Missense |
| 8473 | T>C | ATP8 | syn |
| 8563 | A>G | ATP8 and ATP6 | ATP6: missense;ATP8: syn |
| 8584 | G>A | ATP6 | Missense |
| 8684 | C>T | ATP6 | Missense |
| 8697 | G>A | ATP6 | syn |
| 8701 | A>G | ATP6 | Missense |
| 8705 | T>C | ATP6 | Missense |
| 8727 | C>T | ATP6 | syn |
| 8772 | T>C | ATP6 | syn |
| 8784 | A>G | ATP6 | syn |
| 8793 | T>C | ATP6 | syn |
| 8794 | C>T | ATP6 | Missense |
| 8829 | C>T | ATP6 | syn |
| 8856 | G>A | ATP6 | syn |
| 8860 | A>G | ATP6 | Missense |
| 8943 | C>T | ATP6 | syn |
| 8994 | G>A | ATP6 | syn |
| 9053 | G>A | ATP6 | Missense |
| 9123 | G>A | ATP6 | syn |
| 9128 | T>C | ATP6 | Missense |
| 9180 | A>G | ATP6 | syn |
mtDNA: Mitochondrial DNA; syn: Synonymous; N: Sequence obtained from National Center for Biotechnology Information genbank; T: Mutated sequence in the specimens; A: Adenine; T: Thymine; G: Guanine; C: Cytosine.
The median 2 polymorphic sites (range 1-3) in the MT-ATP 6 and 8 genes were found in control colon specimens (Table 3) while the median 3 polymorphic sites (range 1-4) were found in IBS-D patients (Table 4). Similarly, 1-4 polymorphic sites in MT-ATP 6 and 8 genes from the terminal ileum specimens were found in IBS-D patients (Table 5). When comparing with mtDNA polymorphisms in the colon and terminal ileum, all were identical except in one case (IBS22).
Table 3.
Mitochondrial DNA changes in the colon from controls
| Case |
Nucleotide position |
|||||||||||||||
| 8392 | 8394 | 8414 | 8473 | 8563 | 8584 | 8701 | 8705 | 8727 | 8794 | 8860 | 8994 | 9053 | 9180 | Missense | Sum | |
| 1 | + | + | 2 | 2 | ||||||||||||
| 2 | + | + | + | 2 | 3 | |||||||||||
| 3 | + | + | 2 | 2 | ||||||||||||
| 4 | + | + | 2 | 2 | ||||||||||||
| 5 | + | 1 | 1 | |||||||||||||
| 6 | + | + | 2 | 2 | ||||||||||||
| 7 | + | + | + | 3 | 3 | |||||||||||
| 8 | + | 1 | 1 | |||||||||||||
| 9 | + | + | + | 3 | 3 | |||||||||||
| 10 | + | 1 | 1 | |||||||||||||
| 11 | + | + | 2 | 2 | ||||||||||||
| 12 | + | + | + | 3 | 3 | |||||||||||
| 13 | + | + | 2 | 2 | ||||||||||||
| 14 | + | + | + | 3 | 3 | |||||||||||
| 15 | + | + | 2 | 2 | ||||||||||||
| 16 | + | + | + | 2 | 3 | |||||||||||
| 17 | + | + | 2 | 2 | ||||||||||||
| 18 | + | + | + | 3 | 3 | |||||||||||
| 19 | + | + | + | 2 | 3 | |||||||||||
| 20 | + | + | 2 | 2 | ||||||||||||
| 21 | + | + | + | 3 | 3 | |||||||||||
| 22 | + | + | + | 2 | 3 | |||||||||||
| 23 | + | 1 | 1 | |||||||||||||
| 24 | + | + | 1 | 2 | ||||||||||||
| 25 | + | 1 | 1 | |||||||||||||
| 26 | + | + | + | 2 | 3 | |||||||||||
| 27 | + | + | + | 3 | 3 | |||||||||||
| 28 | + | 1 | 1 | |||||||||||||
Table 4.
Mitochondrial DNA changes in the colon from irritable bowel syndrome group
| Case |
Nucleotide position |
|||||||||||||||||||||
| 8392 | 8414 | 8459 | 8473 | 8563 | 8584 | 8684 | 8697 | 8701 | 8772 | 8784 | 8793 | 8794 | 8829 | 8856 | 8860 | 8943 | 9123 | 9128 | 9180 | Missense | Sum | |
| IBS1 | + | 1 | 1 | |||||||||||||||||||
| IBS2 | + | + | + | 3 | 3 | |||||||||||||||||
| IBS3 | + | + | 1 | 2 | ||||||||||||||||||
| IBS4 | + | 1 | 1 | |||||||||||||||||||
| IBS5 | + | + | + | 3 | 3 | |||||||||||||||||
| IBS6 | + | + | + | 3 | 3 | |||||||||||||||||
| IBS7 | + | + | + | + | 4 | 4 | ||||||||||||||||
| IBS8 | + | + | 1 | 2 | ||||||||||||||||||
| IBS9 | + | + | 1 | 2 | ||||||||||||||||||
| IBS10 | + | + | + | + | 2 | 4 | ||||||||||||||||
| IBS11 | + | + | + | 2 | 3 | |||||||||||||||||
| IBS12 | + | + | + | 2 | 3 | |||||||||||||||||
| IBS13 | + | + | + | + | 3 | 4 | ||||||||||||||||
| IBS14 | + | + | + | + | 3 | 4 | ||||||||||||||||
| IBS15 | + | + | + | + | 2 | 4 | ||||||||||||||||
| IBS16 | + | + | + | + | 2 | 4 | ||||||||||||||||
| IBS17 | + | + | 2 | 2 | ||||||||||||||||||
| IBS18 | + | + | + | + | 3 | 4 | ||||||||||||||||
| IBS19 | + | + | + | 3 | 3 | |||||||||||||||||
| IBS20 | + | + | + | 2 | 3 | |||||||||||||||||
| IBS21 | + | + | 2 | 2 | ||||||||||||||||||
| IBS22 | + | + | 2 | 2 | ||||||||||||||||||
| IBS23 | + | + | + | + | 3 | 4 | ||||||||||||||||
| IBS24 | + | + | + | + | 4 | 4 | ||||||||||||||||
| IBS25 | + | + | 2 | 2 | ||||||||||||||||||
| IBS26 | + | + | 1 | 2 | ||||||||||||||||||
| IBS27 | + | + | 2 | 2 | ||||||||||||||||||
| IBS28 | + | + | + | + | 3 | 4 | ||||||||||||||||
IBS: Irritable bowel syndrome.
Table 5.
Mitochondrial DNA changes in terminal ileum from irritable bowel syndrome group
| Case |
Nucleotide position |
|||||||||||
| 8414 | 8459 | 8473 | 8563 | 8584 | 8684 | 8697 | 8701 | 8772 | 8794 | 8860 | Sum | |
| IBS1 | + | 1 | ||||||||||
| IBS2 | + | + | + | 3 | ||||||||
| IBS3 | + | + | 2 | |||||||||
| IBS5 | + | + | + | 3 | ||||||||
| IBS7 | + | + | + | + | 4 | |||||||
| IBS17 | + | + | 2 | |||||||||
| IBS18 | + | + | + | + | 4 | |||||||
| IBS19 | + | + | + | 3 | ||||||||
| IBS21 | + | + | 2 | |||||||||
| IBS22 | + | + | 2 | |||||||||
IBS: Irritable bowel syndrome.
Association between clinical profiles and mtDNA polymorphisms
Polymorphisms in the MT-ATP 6 and 8 genes were increased in IBS patients as compared with controls (non-parametric Mann-Whitney test, U = 245, P = 0.012; Figure 1). In addition, the difference in percentage of missense polymorphisms between groups was statistically significant (non-parametric Mann-Whitney test, U = 265, P = 0.016). No correlation was found between the presence of mtDNA polymorphisms and disease duration in IBS-D patients (Spearman rho = 0.106, P = 0.592).
Figure 1.
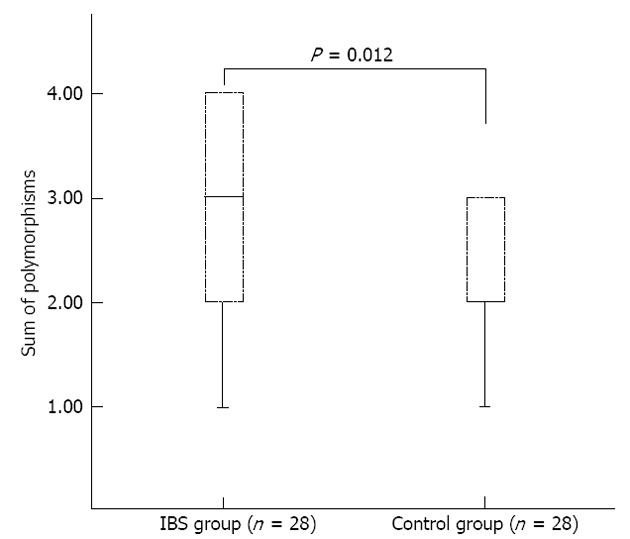
Box plots showing the interquartile range (box) and median (solid line). An increase of mitochondrial DNA polymorphisms in irritable bowel syndrome (IBS) group was observed (P = 0.012).
The mtDNA change at 8860 was present in all cases. The frequency of the 8701 polymorphism was 64% (18/28) in the IBS group and 57% (16/28) in controls, not statistically significant (Pearson χ2 = 0.299, df = 1, P = 0.584). Several IBS-D patients (e.g., IBS13, IBS14 and IBS18) had the same combination of polymorphisms but no similarities in clinical profile (age, gender, duration of disease) were observed.
DISCUSSION
We found that polymorphisms in mitochondrial ATP 6 and 8 genes were more frequent in IBS with diarrhea, than in healthy individuals. Nearly half of the mutations studied were non-synonymous, the meaning of which from a pathophysiological point of view has not been previously clarified.
Blood samples were used typically when analyzing mtDNA. However, previous studies reported that mtDNA changes in the blood were not always the same as those from organ tissues[10,11]. IBS is anatomically attributed to the lower gastrointestinal tract[12]; thus we have chosen colonic and ileal tissues rather than blood. We found that there was almost no difference in mtDNA variants between colon and ileum specimens from the same individual with IBS, except for one case. Both the small and large intestines, harboring the same mtDNA mutation, indicate that a similar pathogenesis may exist in both locations.
mtDNA is highly polymorphic[13,14], and may occur across the entire mtDNA genome, with the MT-ATP gene as one of the hot spots. A high frequency of mtDNA polymorphisms has been implicated in a variety of diseases[15,16], including mitochondrial diseases, degenerative diseases, aging, tumors and so on. Thus far, studies investigating the association between mtDNA and IBS are scarce. Camilleri’s group reported that the mtDNA polymorphism in functional gastrointestinal disorders may be associated with digestive functions, such as satiation, gastric emptying, and pain[7]. Moreover, the mtDNA haplotype might affect the risk of developing IBS. Human mtDNA haplogroups are defined by a particular single nucleotide polymorphism (SNP). Haplotype H (7028C genotype) is predominant in the European population[17,18]; hence, Caucasians are often divided into Haplotype H and non-Haplotype H groups. Camilleri studied the difference between these groups, and found that constipation-predominant and mixed IBS were more likely to be associated with Haplotype H. Most Chinese people are Haplotype A, D and G, but not Haplotype H[19], which is why we did not investigate the haplotype association with IBS in our study.
More than 3000 mtDNA polymorphisms are reported in various mtDNA databases[20], and are often categorized as neutral or pathogenic. The role of pathogenic mtDNA mutations in mitochondrial diseases has been established[21,22], while many polymorphisms in the mtDNA genome present in aging, tumorigenesis and other disease states are considered to be of little pathogenic significance[22,23]. Although efforts have been made to discover the underlying mechanism of these “neutral” polymorphisms, we are far from having a complete understanding of it. Recently, accumulation of mtDNA polymorphisms was proposed to underlie the pathogenesis of these diseases[24-26]. In our study, the polymorphisms detected were neutral, and cumulative polymorphisms in MT-ATP genes were higher in the IBS group. Accumulation of mtDNA polymorphism may play a role in IBS pathogenesis, however, data are limited on functional studies of neutral polymorphisms. In one study, neutral polymorphisms in the mtDNA control region were found to play a role in individual differences such as exercise tolerance[27]. Another study showed that 8701 and 10398 polymorphisms were associated with changes in the mitochondrial matrix pH and intracellular calcium dynamics in the cell line[28], which may help understand the functional significance of some neutral mtDNA polymorphisms. We also identified the 8701 polymorphism, but found no statistical difference in this polymorphism between IBS-D and controls. How these neutral polymorphisms are involved in functional diseases such as IBS-D remains unknown and further research is needed.
This study has several strengths and limitations. A major strength of this study lies in its direct analysis of small intestinal and colon samples, which are considered the areas of IBS symptomatology. As mentioned previously, blood mtDNA represents the status of circulating mitochondria, not specifically that of the bowel. However, due to the nature of intestinal biopsy, it is difficult to recruit a large number of subjects to participate. The relatively small number of study subjects limits our conclusions. Also, there is another concern that no functional studies focusing on these polymorphisms were involved, limiting the interpretation of the results. Further studies with larger sample sizes from multiple centers are required.
In conclusion, we found that cumulative polymorphisms in the mitochondrial ATP genes are associated with IBS-D. While it remains unknown how these polymorphisms are associated with ATP synthase function and cellular energy production in the intestine, our study has important implications for future research on the mtDNA genome in IBS.
ACKNOWLEDGMENTS
We would like to thank Dr. Qing-Sen Liu, Dr. Wen Li and Dr. En-Qiang Linghu for their support. We are also grateful to Drs. Yuan-Zi Yu, and Min Min for their technical assistance. This work was presented in abstract form at UEG Week 2012 in Amsterdam.
COMMENTS
Background
Irritable bowel syndrome (IBS) is one of the most prevalent functional gastrointestinal disorders and affects 10%-20% of the population. To date, the pathogenesis of IBS remains unclear. Family and twin studies suggested that genetic changes may predispose individuals to IBS.
Research frontiers
A previous study indicated that the mitochondrial (MT) haplogroup and variations may be associated with gastrointestinal motor and sensory function in gastrointestinal disorders such as irritable bowel syndrome. Up till now, few research efforts have focused on this topic and little knowledge is available about it.
Innovations and breakthroughs
The authors sequenced the coding region of mitochondrial DNA and found that polymorphisms in MT-ATP 6 and 8 genes were more frequent in IBS with diarrhea, than in healthy individuals.
Applications
This study results implied that the MT-DNA polymorphism may play a role in irritable bowel syndrome with diarrhea, but further studies are needed.
Terminology
MT-DNA: Human MT DNA is approximately 16.6 kbp long, which is inherited solely from the mother and encodes 13 genes for ATP subunits 6 and 8, and other polypeptides of respiratory complexes crucial for ATP production in mitochondria; IBS: According to the Rome III criteria, irritable bowel syndrome is defined as a functional bowel disorder in which abdominal pain or discomfort is associated with defecation or a change in bowel habit, and with features of disordered defecation.
Peer review
In the short paper, the authors analyzed the polymorphisms of MT-ATP 6 and 8 genes in the colon and ileum of IBS with diarrhea (IBS-D). Based on clinical samples, they concluded that patients with IBS-D have a higher incidence of MT-ATP 6 and 8 polymorphisms, compared with healthy controls. This finding is interesting and will be helpful for clinicians to emphasize the importance of the mtDNA polymorphism of IBS-D patients.
Footnotes
P- Reviewers Clave P, Igor SL, Ren XF S- Editor Song XX L- Editor Ma JY E- Editor Li JY
References
- 1.Longstreth GF, Thompson WG, Chey WD, Houghton LA, Mearin F, Spiller RC. Functional bowel disorders. Gastroenterology. 2006;130:1480–1491. doi: 10.1053/j.gastro.2005.11.061. [DOI] [PubMed] [Google Scholar]
- 2.Saito YA, Zimmerman JM, Harmsen WS, De Andrade M, Locke GR, Petersen GM, Talley NJ. Irritable bowel syndrome aggregates strongly in families: a family-based case-control study. Neurogastroenterol Motil. 2008;20:790–797. doi: 10.1111/j.1365-2982.2007.1077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalantar JS, Locke GR, Zinsmeister AR, Beighley CM, Talley NJ. Familial aggregation of irritable bowel syndrome: a prospective study. Gut. 2003;52:1703–1707. doi: 10.1136/gut.52.12.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bengtson MB, Rønning T, Vatn MH, Harris JR. Irritable bowel syndrome in twins: genes and environment. Gut. 2006;55:1754–1759. doi: 10.1136/gut.2006.097287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohammed I, Cherkas LF, Riley SA, Spector TD, Trudgill NJ. Genetic influences in irritable bowel syndrome: a twin study. Am J Gastroenterol. 2005;100:1340–1344. doi: 10.1111/j.1572-0241.2005.41700.x. [DOI] [PubMed] [Google Scholar]
- 6.Saito YA, Mitra N, Mayer EA. Genetic approaches to functional gastrointestinal disorders. Gastroenterology. 2010;138:1276–1285. doi: 10.1053/j.gastro.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camilleri M, Carlson P, Zinsmeister AR, McKinzie S, Busciglio I, Burton D, Zaki EA, Boles RG. Mitochondrial DNA and gastrointestinal motor and sensory functions in health and functional gastrointestinal disorders. Am J Physiol Gastrointest Liver Physiol. 2009;296:G510–G516. doi: 10.1152/ajpgi.90650.2008. [DOI] [PubMed] [Google Scholar]
- 8.Chen XJ, Butow RA. The organization and inheritance of the mitochondrial genome. Nat Rev Genet. 2005;6:815–825. doi: 10.1038/nrg1708. [DOI] [PubMed] [Google Scholar]
- 9.Guo M, Ren J, Brock MV, Herman JG, Carraway HE. Promoter methylation of HIN-1 in the progression to esophageal squamous cancer. Epigenetics. 2008;3:336–341. doi: 10.4161/epi.3.6.7158. [DOI] [PubMed] [Google Scholar]
- 10.Thangaraj K, Joshi MB, Reddy AG, Rasalkar AA, Singh L. Sperm mitochondrial mutations as a cause of low sperm motility. J Androl. 2003;24:388–392. doi: 10.1002/j.1939-4640.2003.tb02687.x. [DOI] [PubMed] [Google Scholar]
- 11.Chong SS, McCall AE, Cota J, Subramony SH, Orr HT, Hughes MR, Zoghbi HY. Gametic and somatic tissue-specific heterogeneity of the expanded SCA1 CAG repeat in spinocerebellar ataxia type 1. Nat Genet. 1995;10:344–350. doi: 10.1038/ng0795-344. [DOI] [PubMed] [Google Scholar]
- 12.Drossman DA. The functional gastrointestinal disorders and the Rome III process. Gastroenterology. 2006;130:1377–1390. doi: 10.1053/j.gastro.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 13.Ingman M, Gyllensten U. mtDB: Human Mitochondrial Genome Database, a resource for population genetics and medical sciences. Nucleic Acids Res. 2006;34:D749–D751. doi: 10.1093/nar/gkj010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brandon MC, Lott MT, Nguyen KC, Spolim S, Navathe SB, Baldi P, Wallace DC. MITOMAP: a human mitochondrial genome database--2004 update. Nucleic Acids Res. 2005;33:D611–D613. doi: 10.1093/nar/gki079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reeve AK, Krishnan KJ, Turnbull D. Mitochondrial DNA mutations in disease, aging, and neurodegeneration. Ann N Y Acad Sci. 2008;1147:21–29. doi: 10.1196/annals.1427.016. [DOI] [PubMed] [Google Scholar]
- 17.Loogväli EL, Roostalu U, Malyarchuk BA, Derenko MV, Kivisild T, Metspalu E, Tambets K, Reidla M, Tolk HV, Parik J, et al. Disuniting uniformity: a pied cladistic canvas of mtDNA haplogroup H in Eurasia. Mol Biol Evol. 2004;21:2012–2021. doi: 10.1093/molbev/msh209. [DOI] [PubMed] [Google Scholar]
- 18.Richards M, Macaulay V, Torroni A, Bandelt HJ. In search of geographical patterns in European mitochondrial DNA. Am J Hum Genet. 2002;71:1168–1174. doi: 10.1086/342930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kivisild T, Tolk HV, Parik J, Wang Y, Papiha SS, Bandelt HJ, Villems R. The emerging limbs and twigs of the East Asian mtDNA tree. Mol Biol Evol. 2002;19:1737–1751. doi: 10.1093/oxfordjournals.molbev.a003996. [DOI] [PubMed] [Google Scholar]
- 20.Bhardwaj A, Mukerji M, Sharma S, Paul J, Gokhale CS, Srivastava AK, Tiwari S. MtSNPscore: a combined evidence approach for assessing cumulative impact of mitochondrial variations in disease. BMC Bioinformatics. 2009;10 Suppl 8:S7. doi: 10.1186/1471-2105-10-S8-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogilvie I, Capaldi RA. Mutation of the mitochrondrially encoded ATPase 6 gene modeled in the ATP synthase of Escherichia coli. FEBS Lett. 1999;453:179–182. doi: 10.1016/s0014-5793(99)00605-5. [DOI] [PubMed] [Google Scholar]
- 22.Wong LJ. Pathogenic mitochondrial DNA mutations in protein-coding genes. Muscle Nerve. 2007;36:279–293. doi: 10.1002/mus.20807. [DOI] [PubMed] [Google Scholar]
- 23.Chatterjee A, Mambo E, Sidransky D. Mitochondrial DNA mutations in human cancer. Oncogene. 2006;25:4663–4674. doi: 10.1038/sj.onc.1209604. [DOI] [PubMed] [Google Scholar]
- 24.Chinnery PF, Samuels DC, Elson J, Turnbull DM. Accumulation of mitochondrial DNA mutations in ageing, cancer, and mitochondrial disease: is there a common mechanism? Lancet. 2002;360:1323–1325. doi: 10.1016/S0140-6736(02)11310-9. [DOI] [PubMed] [Google Scholar]
- 25.Nishikawa M, Oshitani N, Matsumoto T, Nishigami T, Arakawa T, Inoue M. Accumulation of mitochondrial DNA mutation with colorectal carcinogenesis in ulcerative colitis. Br J Cancer. 2005;93:331–337. doi: 10.1038/sj.bjc.6602664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 27.Murakami H, Ota A, Simojo H, Okada M, Ajisaka R, Kuno S. Polymorphisms in control region of mtDNA relates to individual differences in endurance capacity or trainability. Jpn J Physiol. 2002;52:247–256. doi: 10.2170/jjphysiol.52.247. [DOI] [PubMed] [Google Scholar]
- 28.Kazuno AA, Munakata K, Nagai T, Shimozono S, Tanaka M, Yoneda M, Kato N, Miyawaki A, Kato T. Identification of mitochondrial DNA polymorphisms that alter mitochondrial matrix pH and intracellular calcium dynamics. PLoS Genet. 2006;2:e128. doi: 10.1371/journal.pgen.0020128. [DOI] [PMC free article] [PubMed] [Google Scholar]


