Abstract
The POU domain family of transcription factors regulates developmental processes ranging from specification of the early embryo to terminal differentiation. About half of these factors display substantial affinity for an 8 bp DNA site termed the octamer motif, and are hence known as Oct proteins. Oct4 (Pou5f1) is a well-known Oct factor, but there are other Oct proteins with varied and essential roles in development. This Primer outlines our current understanding of Oct proteins and the regulatory mechanisms that govern their role in developmental processes and concludes with the assertion that more investigation into their developmental functions is needed.
Keywords: Oct proteins, POU domain, Stem cells
Introduction
A central tenet of developmental biology is the orchestration of hierarchical gene expression patterns by sequence-specific transcription factors. These gene expression patterns allow for the precise translation of genetic information into the specification of distinct cell lineages, which in turn determines the correct formation of morphological structures. Understanding how this process occurs at a molecular level represents a considerable challenge in developmental biology. Oct transcription factors are key developmental regulators capable of coordinating a spectrum of developmental processes, ranging from the establishment of the embryonic pluripotent ‘ground state’ to terminal differentiation. Oct proteins are a subclass of the POU transcription factor family (Box 1) that recognize an 8 bp consensus sequence [ATGC(A/T)AAT] termed the ‘octamer motif’ and variants thereof (Bodner et al., 1988; Ingraham et al., 1988; Kemler et al., 1989). The 5′ ATGC motif associates with the POU-specific domain, while the 3′ half-site associates with the POU homeodomain (Box 1). The octamer motif is found in the regulatory regions of both ubiquitous and tissue-specific target genes. For example, ubiquitously expressed histone and U small nuclear RNA genes contain the motif in their regulatory regions, but so too do the B cell-restricted immunoglobulin heavy and kappa light chain genes (Fletcher et al., 1987; Henderson and Calame, 1995; Ström et al., 1996).
Box 1. POU domain transcription factors
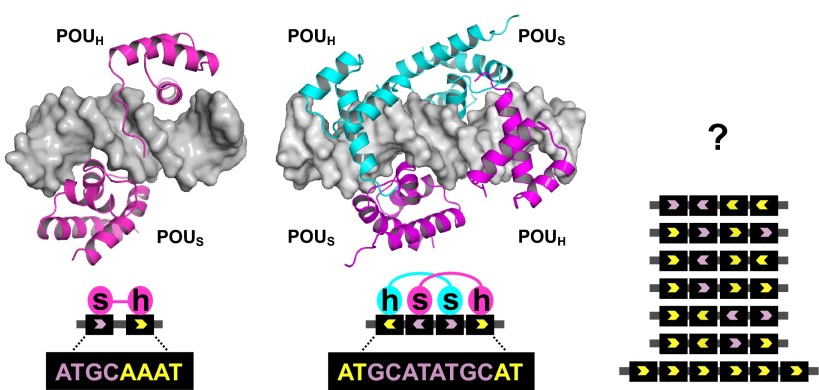
The POU family is a homeodomain subgroup characterized by the presence of a ∼60 amino acid homeodomain (POUH) joined by a flexible linker to a second, independently folded ∼80 amino acid DNA-binding domain termed the POU-specific (POUS) domain (Herr et al., 1988). The two domains make separate contacts with the DNA (Klemm et al., 1994), as illustrated (left) for Oct1 (pink) bound to DNA (gray) at the canonical octamer DNA sequence, the 5′ half of which associates with POUS (s) while the 3′ half associates with POUH (h); in this case, DNA binding is static. Oct proteins can also form dimers or higher-order structures at variants of the canonical binding site, as depicted (center) for an Oct1 homodimer bound to the alternative MORE (more octamer-related palindromic element) sequence (Remenyi et al., 2001), to which binding is inducible by phosphorylation. The mode of multimer (e.g. Oct4) binding to sites identified in vitro by ChIP remains unknown (right), but might be signal responsive in vivo. The combined 150-160 amino acid DNA-binding domain is the hallmark of POU proteins. The strongest single distinguishing POU domain feature lies in the POUH recognition helix: all POU proteins contain the sequence RVWFCN, the sole exception being the atypical POU protein HNF1α (Baumhueter et al., 1990). In non-POU proteins, the cysteine residue in this sequence is typically replaced with a glutamine or serine. The POU DNA-binding structure is grossly similar to a complex of Hoxa9 and a cooperative binding partner, Pbx1, bound to DNA (LaRonde-LeBlanc and Wolberger, 2003). It appears that in the course of POU protein evolution this cooperative association has been solidified by fusing the two ancestral DNA-binding domains together.
POU proteins are expressed in patterns that vary from highly tissue-restricted to ubiquitous. Six POU protein subclasses, termed POU I through POU VI, have been defined based on the sequence composition of the DNA-binding domain. Although POU factors recognize a continuum of related sequences, three classes (POU I, IV and VI) do not display high affinity for the standard octamer motif and so are termed non-octamer-binding factors. The other three classes (POU II, III and V) are termed Oct proteins because they display strong affinity for the octamer motif. As a group, POU proteins tend to be broadly expressed early in development and become progressively more restricted as development proceeds. In the adult, they show highly tissue-restricted expression patterns. An exception to this trend is Oct1, which appears to be the only POU protein that is universally expressed.
Although the basic octamer motif is the simplest sequence capable of strong association with Oct proteins, combinations of paired and inverted octamer half-sites are known to bind proteins such as Oct1 (Pou2f1), Oct4 (Pou5f1, Oct3) and Brn2 (Oct7, N-Oct3, Pou3f2). The proteins bind these sequences as dimers or higher-order structures (Reményi et al., 2001; Nieto et al., 2007; Tantin et al., 2008). The basis of this recognition lies in the conformational flexibility of the linker domain, a covalent peptide of variable length and sequence that allows the two DNA-binding subdomains to reorganize relative to each other (Box 1). The discovery of many diverse Oct protein binding sites mirrors the pattern for other transcription factors, such as Notch intracellular domain, NRSF/REST and T-box transcription factors, in which variant sites with different half-site spacing have been shown to bind the factor physiologically (Conlon et al., 2001; Cave et al., 2005; Ong et al., 2006; Johnson et al., 2007). The capacity for binding to alternative sequences also provides a mechanism for differential regulation by Oct proteins because binding to some sites but not others can be controlled by upstream signals such as phosphorylation (Nieto et al., 2007; Kang et al., 2009).
With these basics in mind, the aim of this Primer is to provide an overview of the known and likely developmental roles of Oct proteins, in particular their relevance to stem cells and how our knowledge of their mechanisms of action can inform their function. Synthesis of prior literature and new mechanistic developments in the field suggests that the time is right to refocus on the roles of these proteins in development. Doing so might result in a renaissance in our understanding of the developmental roles of these key transcription factors.
Oct proteins: a brief overview
Oct proteins comprise POU domain transcription factor classes II, III and V (Table 1). The distinction of these classes is based on sequence similarity of the DNA-binding domain. The best-known Oct protein is the pluripotency regulator Oct4 (Okamoto et al., 1990; Rosner et al., 1990; Schöler et al., 1990), which together with the related protein Sprm1 (Pou5f2) (Pearse et al., 1997), constitutes class V. Oct4 is expressed in the early mammalian embryo and in the germline. Within the early embryo, Oct4 is expressed in pluripotent cells of the blastocyst inner cell mass (ICM) and epiblast, which will eventually give rise to all the cells of the embryo proper. Oct4-deficient embryos fail to establish pluripotency. Instead of forming an ICM, the whole embryo differentiates into trophectoderm and fails to develop following implantation (Nichols et al., 1998). Oct4 is also expressed in embryonic stem cells (ESCs) derived from the ICM, and has been shown to induce reprogramming to ESC-like induced pluripotent stem cells (iPSCs), either alone or in combination with other factors (Takahashi and Yamanaka, 2006; Kim et al., 2009; Zhu et al., 2010).
Table 1.
Mammalian POU transcription factors
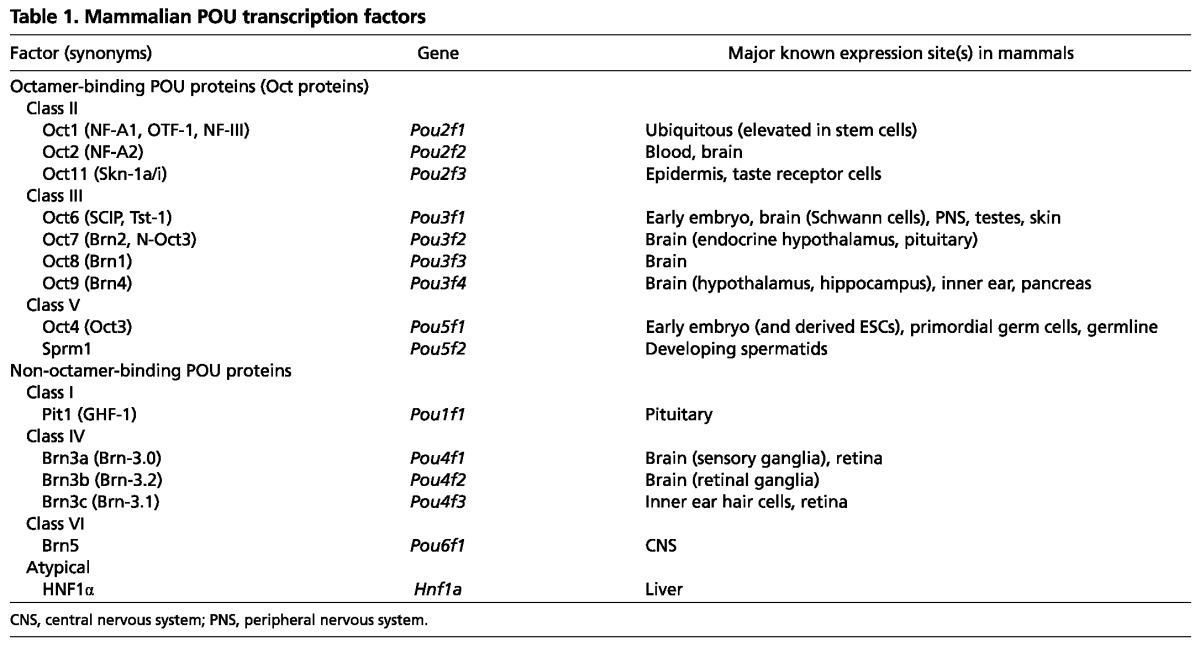
Class II POU domain proteins include prominent Oct proteins such as the ubiquitously expressed Oct1 (Fletcher et al., 1987; Sturm et al., 1987; Sturm et al., 1988) and Oct2 (Pou2f2), which is expressed in the brain and blood (Landolfi et al., 1986; Staudt et al., 1986; Scheidereit et al., 1987; Scheidereit et al., 1988; Staudt et al., 1988; He et al., 1989). Oct1 and Oct2 are the most closely related members, with more than 85% identity in the DNA-binding domain. Another POU class II protein is Oct11 (Skn-1/Pou2f3), which is expressed predominantly in epidermal keratinocytes and taste receptor cells in the taste buds - two cell types that turn over in adult mammals (Andersen et al., 1997; Matsumoto et al., 2011).
Mammalian Oct6 (SCIP, Tst-1, Pou3f1), Brn2, Brn1 (Oct8, Pou3f3) and Brn4 (Oct9, Pou3f4) constitute class III of the POU family. Oct6 is expressed in ESCs, the developing brain and in epidermis (Monuki et al., 1989; Suzuki et al., 1990; Andersen et al., 1997). Brn2 is expressed in specific brain substructures including the neuroendocrine hypothalamus and pituitary (Nakai et al., 1995; Schonemann et al., 1995). Interestingly, Brn2 has been shown to induce direct reprogramming to neuronal lineages when delivered together with other neuronal lineage-specific transcription factors (Pang et al., 2011). Brn2 and Oct4 are thus two POU domain transcription factors with potent lineage reprogramming ability.
The developmental roles of Oct proteins are complex and do not correlate in any obvious way with their degree of homology to one another or with the POU protein class in which they are grouped. Furthermore, it is known that some of these factors, such as Oct4, have unique essential functions whereas others have redundant or compensatory functions that can mask important developmental roles. For example, Brn1 and Brn2 together, but not individually, are crucial for the proper migration of cortical neurons and the proper development of the mammalian neocortex (Sugitani et al., 2002).
Oct protein regulation: interaction partners and signal responsiveness
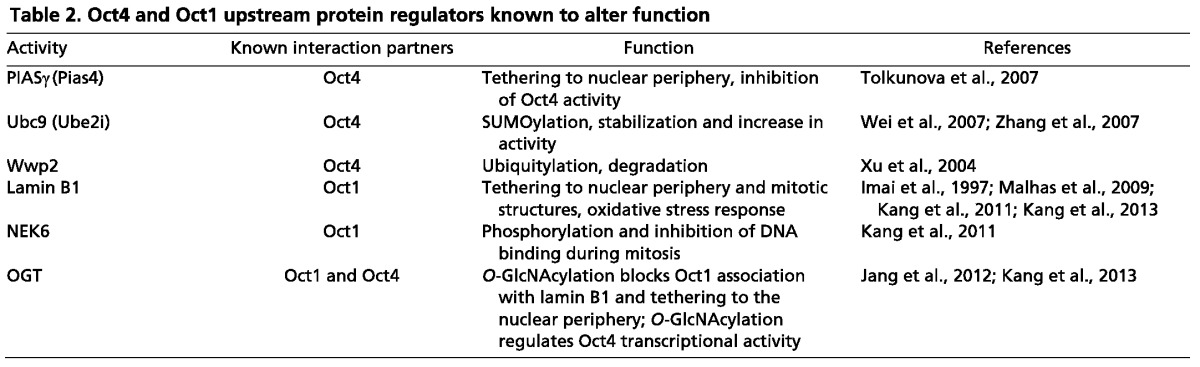
Recent studies have begun to reveal how Oct proteins respond to signals and partner with other proteins to regulate target gene transcription. Oct proteins can integrate multiple upstream signals, which allows precise regulation of their levels, activity and localization. These changes in Oct protein function ensure correct developmental patterning and adult tissue homeostasis. An example of regulated changes in Oct protein levels comes from mammalian peri-implantation embryos. Here, loss of Oct4 is associated with loss of pluripotency and differentiation of epiblast cells (Nichols et al., 1998). Despite the fact that complete loss of Oct4 abolishes pluripotency, slightly attenuated Oct4 levels have been shown to enhance pluripotency in ESCs (Karwacki-Neisius et al., 2013). Thus, the level of Oct4 protein can exert very fine control over the pluripotent state. Oct protein activity can also be regulated by upstream signals, as seen for example with Oct1 and T lymphocytes. Oct1 has been shown to repress the gene that encodes interleukin 2 (Il2) in naïve T cells but switches to an activating mode upon T cell activation (Shakya et al., 2011). This property may allow a single transcription factor to participate at many levels in multistep developmental processes. Control of Oct protein localization is evidenced by the interactions of both Oct4 and Oct1 with their respective upstream regulators (Table 2). Oct4 can be reversibly tethered to the nuclear periphery through interactions with PIASγ (Pias4) (Tolkunova et al., 2007). Oct1 can similarly reversibly associate with the nuclear periphery through interactions with lamin B1. In this latter case association is known to be regulated by oxidative stress, chronic overgrowth and cellular aging (Imai et al., 1997; Malhas et al., 2009; Kang et al., 2013). Other Oct proteins have not been afforded this level of scrutiny, but their functions are likely to be regulated at multiple levels as well.
Table 2.
Oct4 and Oct1 upstream protein regulators known to alter function
Many, possibly all, Oct proteins are subject to regulation through post-translational modifications. These modifications are likely to account for many of the changes in levels, activity and localization described above. The two best-studied Oct proteins in this respect are again Oct4 and Oct1, which are regulated by phosphorylation (Segil et al., 1991; Nieto et al., 2007; Schild-Poulter et al., 2007; Kang et al., 2009; Van Hoof et al., 2009; Kang et al., 2011; Brumbaugh et al., 2012; Lin et al., 2012), O-GlcNAcylation (Webster et al., 2009; Jang et al., 2012; Kang et al., 2013), SUMOylation (Wei et al., 2007; Zhang et al., 2007) and ubiquitylation (Xu et al., 2004; Kang et al., 2011; Kang et al., 2013). These modifications control protein stability, their DNA binding properties, co-factor association and tethering to the nuclear envelope. This array of modifications is unlikely to produce an on/off-type response, but rather a continuum of different outputs.
Developmentally important transcription factors can be regulated via changes in the levels or activity of partner proteins with which they form cooperative (or sometimes antagonistic) DNA-bound complexes. The ability to form these cooperative complexes alters and expands the DNA sequence recognition space available to individual factors. This paradigm applies in the case of Oct proteins, although here the case is more unusual because POU proteins are already in effect cooperative DNA-binding ‘complexes’ by virtue of their two independently folded DNA-binding subdomains (Box 1). Both Oct1 and Oct4 associate with Sox2 at DNA elements composed of ‘Oct/Sox’ composite binding sites (Ambrosetti et al., 1997). The requirement for Sox2 in ESCs can be eliminated by slightly elevating Oct4 expression, indicating a fine balance of activities necessary to maintain pluripotency (Masui et al., 2007). Recent findings suggest that Oct4 can also bind DNA cooperatively with the related protein Sox17 at slightly different composite elements with shorter spacing between the DNA binding sites. These alternative sites are found in genes that specify endoderm (Aksoy et al., 2013).
Oct4 has been shown to additionally associate with Klf4 (Wei et al., 2009), ESRRB (van den Berg et al., 2008), Nanog (Liang et al., 2008) and Zfp143 (Chen et al., 2008a) at different binding sites. Oct1 has been shown to associate not only with transcription factors such as Sox2, C/EBPβ, AP-1 (Fos), NFAT, NFκB and glucocorticoid receptor (Li-Weber et al., 1998; Préfontaine et al., 1999; Belsham and Mellon, 2000; Hatada et al., 2000; van Heel et al., 2002), but also with itself and with other Oct proteins such as Oct2 and Oct6 to form homodimeric, heterodimeric and higher-order complexes (Verrijzer et al., 1992; Reményi et al., 2001). Interestingly, the ability to form these complexes can be controlled by phosphorylation of Oct proteins at specific sites in the DNA-binding domain (Nieto et al., 2007; Kang et al., 2009). Co-incident Oct1 and Oct4 binding has been described for a specific subset of Oct4 targets in ESCs. In some cases, sequential chromatin immunoprecipitation (ChIP) indicates that binding occurs simultaneously at the same DNA site (Ferraris et al., 2011). These binding events might therefore be sites of Oct1/Oct4 heterodimer or higher-order complex formation. Much more work will be required to fully understand the interplay between different Oct proteins at their binding sites in different tissues and developmental stages.
Table 3 shows some of the proteins known to mediate the downstream functions of Oct4 or Oct1. These co-factors can act positively, combining with the Oct protein to activate gene expression, or alternatively they may have inhibitory effects. For example, general transcription factors may play an activating role when the Oct protein-binding site in the DNA is exceptionally close to the core promoter, as has been shown for Oct1 (Nakshatri et al., 1995; Inamoto et al., 1997; Bertolino and Singh, 2002). Paf1, Wdr5 and XPC have all been identified as positive co-factors, interacting with Oct4 to maintain pluripotency in ESCs (Ponnusamy et al., 2009; Ang et al., 2011; Fong et al., 2011). Although the mechanisms are not always clear, these co-factors colocalize with Oct4 at specific target genes and help Oct4 mediate its positive transcriptional functions. By contrast, SETDB1 (ESET), a histone H3K9 methyltransferase, associates with Oct4 to repress target gene expression (Yeap et al., 2009; Yuan et al., 2009). NuRD, a multi-subunit complex with nucleosome remodeling and histone deacetylase activity, is another repressive activity known to associate with Oct4 (Liang et al., 2008). Interestingly, three separate studies have identified proteins that physically interact with Oct4 using mass spectroscopy (Pardo et al., 2010; van den Berg et al., 2010; Ding et al., 2012) but only a small number of the proteins (18/263) overlap between the three studies. It is not clear how many of these are direct rather than indirect Oct4 interaction partners, but among the 18 proteins are subunits of NuRD. Oct1 also interacts robustly with NuRD to repress target genes (Shakya et al., 2011). When Oct1 changes to an activating mode, it associates with KDM3A (Jmjd1a), a histone lysine demethylase that removes inhibitory histone marks (Shakya et al., 2011). Upstream signaling through the MEK/ERK pathway mediates Oct1 switching between NuRD and KDM3A to regulate the expression of target genes such as Il2 and Cdx2 (Shakya et al., 2011).
Table 3.
Mediators of downstream Oct protein activity
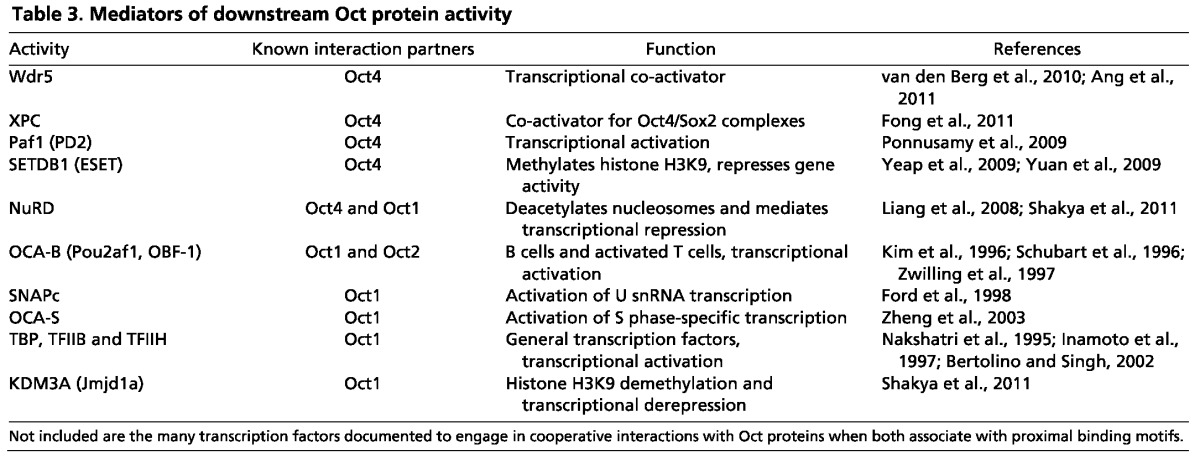
Despite the identification of numerous Oct-interacting proteins, there are still fundamental unresolved questions concerning the mechanisms that underlie the differential utilization of Oct co-factors and regulators. Most of the work in this area has involved Oct4 or Oct1, and it is not clear which of these mechanisms can be generalized to all or part of the Oct protein family. Crucially, with the exception of the Oct1 example provided above, it is still unclear how the same Oct protein is able to switch between activating and repressive states. Post-translational modifications, as well as the specific complexes formed between the Oct protein and other DNA-binding proteins, may determine the recruited effector protein(s) and thus the transcriptional output.
Modes of transcriptional regulation and gene poising by Oct proteins
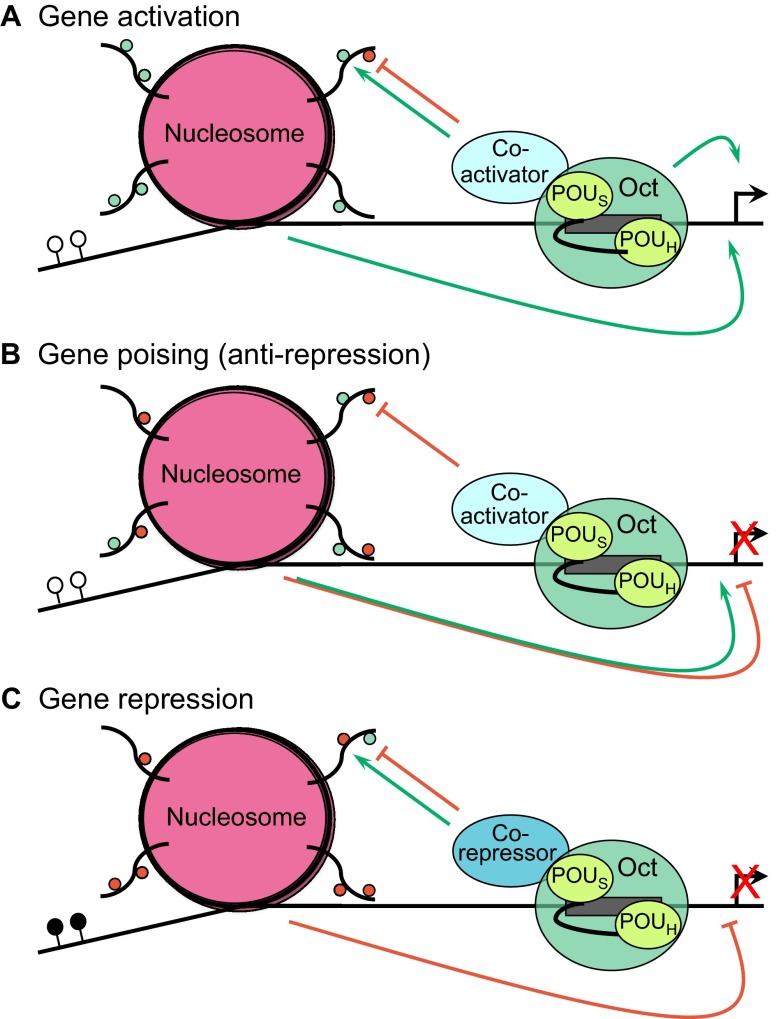
Oct targets can be placed into three broad categories: (1) positive targets that are actively expressed; (2) positive targets that are transcriptionally silent; and (3) negative (repressed) targets (Fig. 1). For those targets in category 2, the Oct protein or proteins maintain epigenetically ‘poised’ gene expression states: situations in which a target gene is silent but readily inducible upon reception of the appropriate developmental cues.
Fig. 1.
Modes of Oct protein transcriptional regulation. The binding of Oct protein (green) to a target gene depends on the presence of a sequence-specific binding site (gray rectangle) and involves the POUH and POUS domains (light green) within the Oct protein, which are connected via a linker sequence (see Box 1). (A) Gene activation. The Oct protein activates the expression of its target genes (green arrows) either directly by promoting the assembly of a transcription complex at the promoter or indirectly by acting through the nucleosomes (pink). In the latter case, the Oct protein interacts with co-factors (blue) to deposit positively acting chromatin modifications (small green circles) or to remove repressive marks (small red circles). ATP-dependent chromatin remodeling may also be employed (not shown). Active gene expression (black arrow on the DNA) is typically associated with promoter DNA that is free of cytosine methylation (white circles). (B) Poising of transcriptionally silent genes. Targets genes of Oct1 and Oct4, and perhaps other Oct proteins, can be transcriptionally silent (red X) but epigenetically poised to rapidly initiate expression upon reception of the correct developmental cues. Present evidence indicates that the Oct protein mechanism in gene poising is positive, removing repressive modifications. The effect of chromatin on gene expression can be differentiated from that in A because a mixture of positively and negatively acting modifications is present (simultaneous green and red arrows). These genes typically lack DNA methylation (white circles). (C) Gene repression. In this case, the Oct protein recruits co-repressor activities that remove activating marks or deposit repressive marks on the chromatin to repress gene transcription (red arrow). These genes are typified by a high degree of promoter DNA methylation (black circles).
Oct4 provides examples of all three classes. Category 1 Oct4 targets in ESCs include active pluripotency genes such as Nanog. Oct4 also positively regulates its own gene (Pou5f1). Nanog, Oct4 and a small number of other proteins form what is termed the ‘core pluripotency network’, which is required to maintain pluripotency. Their expression is maintained not only by Oct4 but also by other proteins encoded in the network such as Nanog and Sox2 (Boyer et al., 2005; Rodda et al., 2005). In addition to the core pluripotency network, a larger number of additional category 1 Oct4 targets encode constitutively expressed proteins such as histones and metabolic enzymes (Chen et al., 2008b). Category 3 Oct4 targets include Cdx2, which is actively repressed by Oct4 and is silent in ESCs (Yeap et al., 2009; Yuan et al., 2009). Finally, hundreds of developmentally poised category 2 Oct4 targets are silent in ESCs but become activated in different developing tissues, e.g. Hoxa5, Hoxc6, Pax6, Otx2, Gata2 and Pou4f1 (Brn3a) (Bernstein et al., 2006; Chen et al., 2008b). They often encode tissue-specific, lineage-instructive transcription factors. In ESCs, category 2 genes are characterized by a chromatin state free of DNA methylation and containing both activating and repressive histone marks (Bernstein et al., 2006; Meissner et al., 2008). These genes are rapidly induced if differentiating cells encounter the appropriate developmental cues, or become stably repressed if cells proceed down other developmental trajectories. Oct4 is not only expressed in ESCs but also in primordial germ cells and spermatogonial stem cells (Rosner et al., 1990; Kehler et al., 2004). It is possible that Oct4 associates with the same category 2 genes in these cells as well, and thus the activity of Oct4 might be a unifying feature of totipotent/pluripotent stem cells of both the germline and embryo. It is difficult to test mechanistically the poising function of Oct4 in ESCs because of its simultaneous role in maintaining the pluripotency network. Ablating Oct4 in ESCs causes the network to collapse and ESCs to differentiate. Nevertheless, the strong correlation between Oct4 binding to these targets and their poised configuration strongly suggests that Oct4 is executing a poising function. These findings regarding Oct4 target loci in ESCs highlight the importance of distinguishing between silent repressed targets (in which active repression is taking place) and silent poised targets (in which the molecular action of the Oct protein at the target is presumably positive, Fig. 1). Simple evaluation of target gene expression levels is inadequate for this task. Instead, the presence of particular co-factors and the status of local chromatin (Fig. 1) at the target need to be determined.
Other Oct protein family members also mediate different target gene expression states. Oct1 has been directly implicated in gene poising at the Il2 locus in resting but previously stimulated CD4 T cells (Shakya et al., 2011). In this study it was found that, within 6 hours of naïve CD4 T cell stimulation, Oct1 switches Il2 from a repressive (category 1) to a poising (category 2) state. Switching was dependent on MAP kinase signals, indicating that Oct proteins can rapidly switch the manner in which they regulate target genes in response to upstream signals. After withdrawal of the initial stimulus and attenuation of Il2 expression, Oct1 blocked the complete repression of this locus, maintaining it in a poised state such that the cells responded more quickly and with greatly augmented Il2 expression upon reactivation (Shakya et al., 2011). This ‘anti-repression’ is achieved via an interaction of Oct1 with the chromatin-modifying protein KDM3A, which opposes inhibitory histone methylation of the Il2 promoter.
Mammalian ESCs co-express Oct4 with Oct1 and Oct6 (Okamoto et al., 1990; Rosner et al., 1990; Schöler, 1991). Oct1 co-occupies many of the same poised developmental Oct4 targets but not the active pluripotency Oct4 targets (Ferraris et al., 2011). Oct6 binding to endogenous target genes has not been tested. It is unclear whether Oct1 or Oct6 is also involved in poising developmental genes in ESCs and pluripotent cells of the embryo.
Indirect evidence implicates three other mammalian Oct proteins, Oct2, Brn2 and Brn4, in gene poising. Although Oct2 is increasingly expressed throughout adult B cell development, Oct2-deficient B cells mature normally. Instead, mature B cells manifest multiple defects upon activation, including poor proliferation and poor terminal differentiation to plasma cells (Corcoran et al., 1993; Corcoran and Karvelas, 1994; Emslie et al., 2008). These results are consistent with a gene poising role of Oct2 during B cell development in a manner that might be conceptually and mechanistically similar to Oct1, its closest paralog. Similarly, the POU III Oct protein Brn2 is expressed throughout mammalian neuroendocrine hypothalamic development, but in the null condition defects only manifest during terminal differentiation (Nakai et al., 1995; Schonemann et al., 1995). It is therefore possible that Brn2 is also involved in poising specific targets at the earlier developmental stages, with the phenotype manifesting only when critical target genes need to be properly induced. In the developing brain, another POU III Oct protein, Brn4, is widely expressed in the ventricular zone, including in striatal stem and progenitor cells. However, depletion of Brn4 specifically reduces the number of differentiated neurons (Shimazaki et al., 1999). These findings are also consistent with a model in which Brn4 poises key targets at earlier developmental stages. These possibilities await formal testing through assessment of the chromatin at target genes and careful measurement of gene induction following manipulation of these activities. In summary, it is possible that many Oct proteins mediate poised chromatin states at subsets of their targets.
Oct proteins and somatic stem cells
Pluripotent and somatic stem cells are critically dependent on poised gene expression states for proper function. Stem cells also frequently (but not uniformly) show properties such as a glycolytic metabolic signature, relative resistance to oxidative and genotoxic stress, and the ability to divide asymmetrically. Interestingly, both Oct4 and Oct1 regulate target genes involved in core carbon metabolism (Chen et al., 2008b; Shakya et al., 2009). Higher levels of Oct1 promote both stress resistance (Tantin et al., 2005) and a glycolytic metabolic profile associated with cellular transformation (Shakya et al., 2009). The metabolic and stress response phenotypes suggest that Oct1 might regulate functions associated with stem cells. Recent results show that Oct1 is highly expressed in the stem cell compartments of the gastrointestinal and blood systems and that Oct1 promotes multiple phenotypes associated with stem cell function, such as hematopoietic engraftment ability (Maddox et al., 2012). These results strongly suggest that Oct1 regulates stem cell function in a range of tissue-specific contexts.
Apart from Oct1 and Oct4, there is tantalizing but tangential evidence that other Oct proteins regulate the stem cell phenotype. As mentioned above, the mammalian Oct2, Brn2 and Brn4 loss-of-function phenotypes are consistent with a role in gene poising in progenitor/stem cell compartments. Other evidence comes from non-mouse models. A summary of POU proteins in three common non-mammalian model organisms, Drosophila melanogaster, Caenorhabditis elegans and Danio rerio, is shown in Table 4. There are five Drosophila POU proteins (Table 4). Two of these, Pdm1 (Nubbin - FlyBase) and Pdm2, are Oct proteins belonging to POU class II, the same group in which mammalian Oct1, Oct2 and Oct11 are found. Pdm1 and Pdm2 are required to maintain self-renewing asymmetric divisions in neuronal progenitor cells and therefore to generate post-mitotic daughter neurons (Bhat and Apsel, 2004). Interestingly, a specifically phosphorylated form of mammalian Oct1 localizes to mitotic structures, such as spindle pole bodies and the midbody, in symmetrically dividing HeLa and fibroblast cells (Kang et al., 2011). The mechanism underlying this function of Pdm1/2, and the potential role of Oct1 modification in asymmetrically dividing cells, have not been tested.
Table 4.
POU domain proteins in three common model organisms
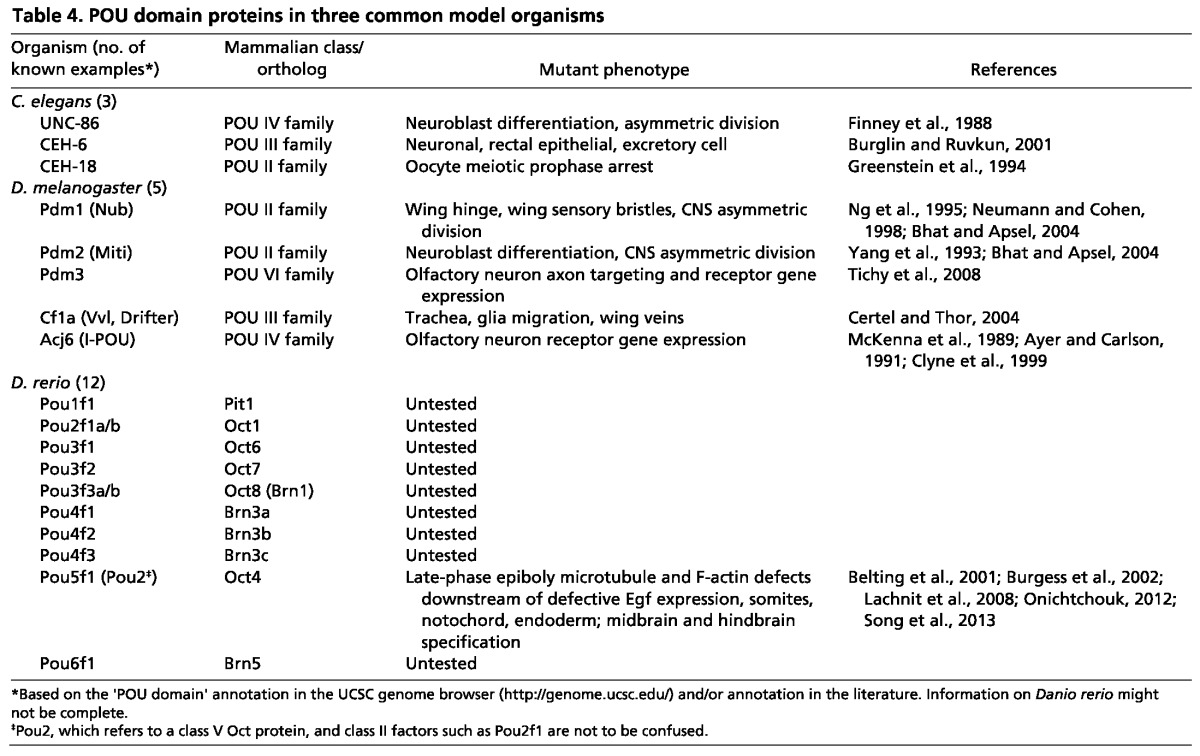
Relatively little is known about the role of zebrafish Oct proteins in stem cells and development (Box 2). The best studied is Pou5f1 (Pou2, Pou5f3), which shares similarities with mammalian Oct4, although both the expression patterns and development defects associated with its inactivation suggest a different mechanism and function for Pou5f1. Further studies of zebrafish Oct proteins are needed to better understand their diverse mechanisms and functions. In addition to those listed in Table 4, there are published studies of Oct proteins from a number of other organisms (e.g. Xenopus, fugu, medaka, axotoxl, opossum). Most of this work involves comparative analysis of Oct4 orthologs and is beyond the scope of this Primer. Work on other Oct proteins in these organisms largely awaits investigation.
Box 2. Zebrafish Oct proteins
The zebrafish genome encodes at least ten non-redundant POU domain-containing factors, five of which are clear Oct protein orthologs (Table 4). The roles of most of these are poorly characterized, although Pou5f1 (Pou2) is the best studied (Belting et al., 2001; Burgess et al., 2002; Lachnit et al., 2008; Onichtchouk, 2012; Song et al., 2013). Historically, the term POU2 has been applied to numerous POU class V Oct4 orthologs in different vertebrates, including zebrafish. These Oct4-like class V POU2 factors should not be confused with POU class II transcription factors. Although zebrafish Pou5f1 and mammalian Oct4 are related to each other and both knockouts result in developmental defects, their expression patterns are different and so the two proteins cannot be directly equated. Fish deficient in maternal zygotic Pou5f1 display pleiotropic phenotypes around the time of gastrulation. These include overall dorsalization of the embryo, defective Egf expression leading to cytoskeletal defects and delay of epiboly, and deficient endoderm formation (Burgess et al., 2002; Lachnit et al., 2008; Onichtchouk, 2012; Song et al., 2013). In addition, in the developing brain Pou5f1 ablation results in severe defects in the midbrain and hindbrain primordia (Belting et al., 2001; Burgess et al., 2002). This complex phenotype suggests that the single zebrafish Pou5f1 activity executes the functions of multiple mammalian paralogs, some of which are unlikely to be specific to stem cells. Among vertebrates, zebrafish Pou5f1 might be somewhat of an outlier in this regard. Whereas zebrafish Pou5f1 cannot reprogram mammalian fibroblasts to pluripotency, medaka and axotoxl Pou5f1 can (Tapia et al., 2012). These findings suggest that this function is ancestral and has been lost in zebrafish.
Not all Oct proteins are likely to promote stem cell phenotypes. Some Oct proteins could antagonize stem cell phenotypes by opposing other Oct proteins, for example by competitive binding and/or through differential transcriptional regulatory mechanisms. One interesting example comes from mammalian brain development and POU class III Oct proteins. In neural progenitor cells of the forebrain and midbrain the members of this class - Oct6, Brn2, Brn1 and Brn4 - all associate with an Otx2 upstream enhancer. By contrast, an antagonistic transcription factor, Gbx2, associates and represses this locus in hindbrain (Inoue et al., 2012). These results reveal a complex interplay between multiple Oct proteins and other factors at a single enhancer binding site. Also, different isoforms expressed from the same gene locus can oppose other isoforms; for example, different Oct2 isoforms appear to play opposing roles in the neuronal differentiation of ESCs (Theodorou et al., 2009).
Oct proteins in development and stem cells: an interim conclusion
Oct protein research should be spurred on by the impressive findings of the last 10 years regarding Oct4. Nevertheless, many unanswered questions remain regarding their mechanisms of action and developmental functions. Importantly, the great majority of zebrafish Oct proteins remain completely uncharacterized. These issues are best addressed by leveraging new molecular tools that enable a better understanding of the target genes and regulatory mechanisms utilized by Oct proteins. Powerful new methods for ablating specific genes in zebrafish can be applied to manipulate Oct proteins. For many of the mammalian family members, conditional knockout alleles have not been constructed. The ability to ablate the activity of different factors in a given cell type and in a temporally controlled fashion will help to further elucidate the developmental roles of these important transcription factors.
Based on the prior work in this field, the results of future investigations are likely to be complex but also enlightening. One specific area of focus will be gene poising by Oct proteins and how this mechanism enables their different roles in pluripotent stem cell, somatic stem cell and progenitor cell compartments. The fact that Oct proteins are implicated in gene poising could complicate the interpretation of their function during development. A specific expression or DNA binding pattern may only partially indicate function, as the protein might be poising critical target genes that become activated at later developmental stages. Therefore, investigations of developmental phenotypes associated with Oct proteins in different model organisms and developmental paradigms must go hand-in-hand with advances in understanding the mechanisms by which these proteins function.
Acknowledgments
We thank R. Dorsky, D. Stillman and members of the D.T. laboratory for their critical reading of the manuscript. We apologize to authors whose work was omitted from this article due to space limitations.
Footnotes
Funding
Work in the D.T. laboratory was supported by the National Institutes of Health and by an award from the Concern Foundation. Deposited in PMC for release after 12 months.
Competing interests statement
The author declares no competing financial interests.
References
- Aksoy I., Jauch R., Chen J., Dyla M., Divakar U., Bogu G. K., Teo R., Leng Ng C. K., Herath W., Lili S., et al. (2013). Oct4 switches partnering from Sox2 to Sox17 to reinterpret the enhancer code and specify endoderm. EMBO J. 32, 938–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosetti D. C., Basilico C., Dailey L. (1997). Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol. Cell. Biol. 17, 6321–6329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen B., Weinberg W. C., Rennekampff O., McEvilly R. J., Bermingham J. R., Jr, Hooshmand F., Vasilyev V., Hansbrough J. F., Pittelkow M. R., Yuspa S. H., et al. (1997). Functions of the POU domain genes Skn-1a/i and Tst-1/Oct-6/SCIP in epidermal differentiation. Genes Dev. 11, 1873–1884 [DOI] [PubMed] [Google Scholar]
- Ang Y. S., Tsai S. Y., Lee D. F., Monk J., Su J., Ratnakumar K., Ding J., Ge Y., Darr H., Chang B., et al. (2011). Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 145, 183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayer R. K., Jr, Carlson J. (1991). acj6: a gene affecting olfactory physiology and behavior in Drosophila. Proc. Natl. Acad. Sci. USA 88, 5467–5471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumhueter S., Mendel D. B., Conley P. B., Kuo C. J., Turk C., Graves M. K., Edwards C. A., Courtois G., Crabtree G. R. (1990). HNF-1 shares three sequence motifs with the POU domain proteins and is identical to LF-B1 and APF. Genes Dev. 4, 372–379 [DOI] [PubMed] [Google Scholar]
- Belsham D. D., Mellon P. L. (2000). Transcription factors Oct-1 and C/EBPbeta (CCAAT/enhancer-binding protein-beta) are involved in the glutamate/nitric oxide/cyclic-guanosine 5′-monophosphate-mediated repression of mediated repression of gonadotropin-releasing hormone gene expression. Mol. Endocrinol. 14, 212–228 [DOI] [PubMed] [Google Scholar]
- Belting H. G., Hauptmann G., Meyer D., Abdelilah-Seyfried S., Chitnis A., Eschbach C., Söll I., Thisse C., Thisse B., Artinger K. B., et al. (2001). spiel ohne grenzen/pou2 is required during establishment of the zebrafish midbrain-hindbrain boundary organizer. Development 128, 4165–4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 [DOI] [PubMed] [Google Scholar]
- Bertolino E., Singh H. (2002). POU/TBP cooperativity: a mechanism for enhancer action from a distance. Mol. Cell 10, 397–407 [DOI] [PubMed] [Google Scholar]
- Bhat K. M., Apsel N. (2004). Upregulation of Mitimere and Nubbin acts through cyclin E to confer self-renewing asymmetric division potential to neural precursor cells. Development 131, 1123–1134 [DOI] [PubMed] [Google Scholar]
- Bodner M., Castrillo J. L., Theill L. E., Deerinck T., Ellisman M., Karin M. (1988). The pituitary-specific transcription factor GHF-1 is a homeobox-containing protein. Cell 55, 505–518 [DOI] [PubMed] [Google Scholar]
- Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., et al. (2005). Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbaugh J., Hou Z., Russell J. D., Howden S. E., Yu P., Ledvina A. R., Coon J. J., Thomson J. A. (2012). Phosphorylation regulates human OCT4. Proc. Natl. Acad. Sci. USA 109, 7162–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S., Reim G., Chen W., Hopkins N., Brand M. (2002). The zebrafish spiel-ohne-grenzen (spg) gene encodes the POU domain protein Pou2 related to mammalian Oct4 and is essential for formation of the midbrain and hindbrain, and for pre-gastrula morphogenesis. Development 129, 905–916 [DOI] [PubMed] [Google Scholar]
- Bürglin T. R., Ruvkun G. (2001). Regulation of ectodermal and excretory function by the C. elegans POU homeobox gene ceh-6. Development 128, 779–790 [DOI] [PubMed] [Google Scholar]
- Cave J. W., Loh F., Surpris J. W., Xia L., Caudy M. A. (2005). A DNA transcription code for cell-specific gene activation by notch signaling. Curr. Biol. 15, 94–104 [DOI] [PubMed] [Google Scholar]
- Certel S. J., Thor S. (2004). Specification of Drosophila motoneuron identity by the combinatorial action of POU and LIM-HD factors. Development 131, 5429–5439 [DOI] [PubMed] [Google Scholar]
- Chen X., Fang F., Liou Y. C., Ng H. H. (2008a). Zfp143 regulates Nanog through modulation of Oct4 binding. Stem Cells 26, 2759–2767 [DOI] [PubMed] [Google Scholar]
- Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V. B., Wong E., Orlov Y. L., Zhang W., Jiang J., et al. (2008b). Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 [DOI] [PubMed] [Google Scholar]
- Clyne P. J., Certel S. J., de Bruyne M., Zaslavsky L., Johnson W. A., Carlson J. R. (1999). The odor specificities of a subset of olfactory receptor neurons are governed by Acj6, a POU-domain transcription factor. Neuron 22, 339–347 [DOI] [PubMed] [Google Scholar]
- Conlon F. L., Fairclough L., Price B. M., Casey E. S., Smith J. C. (2001). Determinants of T box protein specificity. Development 128, 3749–3758 [DOI] [PubMed] [Google Scholar]
- Corcoran L. M., Karvelas M. (1994). Oct-2 is required early in T cell-independent B cell activation for G1 progression and for proliferation. Immunity 1, 635–645 [DOI] [PubMed] [Google Scholar]
- Corcoran L. M., Karvelas M., Nossal G. J., Ye Z. S., Jacks T., Baltimore D. (1993). Oct-2, although not required for early B-cell development, is critical for later B-cell maturation and for postnatal survival. Genes Dev. 7, 570–582 [DOI] [PubMed] [Google Scholar]
- Ding J., Xu H., Faiola F., Ma’ayan A., Wang J. (2012). Oct4 links multiple epigenetic pathways to the pluripotency network. Cell Res. 22, 155–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emslie D., D’Costa K., Hasbold J., Metcalf D., Takatsu K., Hodgkin P. O., Corcoran L. M. (2008). Oct2 enhances antibody-secreting cell differentiation through regulation of IL-5 receptor alpha chain expression on activated B cells. J. Exp. Med. 205, 409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraris L., Stewart A. P., Kang J., DeSimone A. M., Gemberling M., Tantin D., Fairbrother W. G. (2011). Combinatorial binding of transcription factors in the pluripotency control regions of the genome. Genome Res. 21, 1055–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney M., Ruvkun G., Horvitz H. R. (1988). The C. elegans cell lineage and differentiation gene unc-86 encodes a protein with a homeodomain and extended similarity to transcription factors. Cell 55, 757–769 [DOI] [PubMed] [Google Scholar]
- Fletcher C., Heintz N., Roeder R. G. (1987). Purification and characterization of OTF-1, a transcription factor regulating cell cycle expression of a human histone H2b gene. Cell 51, 773–781 [DOI] [PubMed] [Google Scholar]
- Fong Y. W., Inouye C., Yamaguchi T., Cattoglio C., Grubisic I., Tjian R. (2011). A DNA repair complex functions as an Oct4/Sox2 coactivator in embryonic stem cells. Cell 147, 120–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford E., Strubin M., Hernandez N. (1998). The Oct-1 POU domain activates snRNA gene transcription by contacting a region in the SNAPc largest subunit that bears sequence similarities to the Oct-1 coactivator OBF-1. Genes Dev. 12, 3528–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenstein D., Hird S., Plasterk R. H., Andachi Y., Kohara Y., Wang B., Finney M., Ruvkun G. (1994). Targeted mutations in the Caenorhabditis elegans POU homeo box gene ceh-18 cause defects in oocyte cell cycle arrest, gonad migration, and epidermal differentiation. Genes Dev. 8, 1935–1948 [DOI] [PubMed] [Google Scholar]
- Hatada E. N., Chen-Kiang S., Scheidereit C. (2000). Interaction and functional interference of C/EBPbeta with octamer factors in immunoglobulin gene transcription. Eur. J. Immunol. 30, 174–184 [DOI] [PubMed] [Google Scholar]
- He X., Treacy M. N., Simmons D. M., Ingraham H. A., Swanson L. W., Rosenfeld M. G. (1989). Expression of a large family of POU-domain regulatory genes in mammalian brain development. Nature 340, 35–41 [DOI] [PubMed] [Google Scholar]
- Henderson A. J., Calame K. L. (1995). Lessons in transcriptional regulation learned from studies on immunoglobulin genes. Crit. Rev. Eukaryot. Gene Expr. 5, 255–280 [DOI] [PubMed] [Google Scholar]
- Herr W., Sturm R. A., Clerc R. G., Corcoran L. M., Baltimore D., Sharp P. A., Ingraham H. A., Rosenfeld M. G., Finney M., Ruvkun G., et al. (1988). The POU domain: a large conserved region in the mammalian pit-1, oct-1, oct-2, and Caenorhabditis elegans unc-86 gene products. Genes Dev. 2 12A, 1513–1516 [DOI] [PubMed] [Google Scholar]
- Imai S., Nishibayashi S., Takao K., Tomifuji M., Fujino T., Hasegawa M., Takano T. (1997). Dissociation of Oct-1 from the nuclear peripheral structure induces the cellular aging-associated collagenase gene expression. Mol. Biol. Cell 8, 2407–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamoto S., Segil N., Pan Z. Q., Kimura M., Roeder R. G. (1997). The cyclin-dependent kinase-activating kinase (CAK) assembly factor, MAT1, targets and enhances CAK activity on the POU domains of octamer transcription factors. J. Biol. Chem. 272, 29852–29858 [DOI] [PubMed] [Google Scholar]
- Ingraham H. A., Chen R. P., Mangalam H. J., Elsholtz H. P., Flynn S. E., Lin C. R., Simmons D. M., Swanson L., Rosenfeld M. G. (1988). A tissue-specific transcription factor containing a homeodomain specifies a pituitary phenotype. Cell 55, 519–529 [DOI] [PubMed] [Google Scholar]
- Inoue F., Kurokawa D., Takahashi M., Aizawa S. (2012). Gbx2 directly restricts Otx2 expression to forebrain and midbrain, competing with class III POU factors. Mol. Cell. Biol. 32, 2618–2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H., Kim T. W., Yoon S., Choi S. Y., Kang T. W., Kim S. Y., Kwon Y. W., Cho E. J., Youn H. D. (2012). O-GlcNAc regulates pluripotency and reprogramming by directly acting on core components of the pluripotency network. Cell Stem Cell 11, 62–74 [DOI] [PubMed] [Google Scholar]
- Johnson D. S., Mortazavi A., Myers R. M., Wold B. (2007). Genome-wide mapping of in vivo protein-DNA interactions. Science 316, 1497–1502 [DOI] [PubMed] [Google Scholar]
- Kang J., Gemberling M., Nakamura M., Whitby F. G., Handa H., Fairbrother W. G., Tantin D. (2009). A general mechanism for transcription regulation by Oct1 and Oct4 in response to genotoxic and oxidative stress. Genes Dev. 23, 208–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Goodman B., Zheng Y., Tantin D. (2011). Dynamic regulation of Oct1 during mitosis by phosphorylation and ubiquitination. PLoS ONE 6, e23872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Shen Z., Lim J.-M., Handa H., Wells L., Tantin D. (2013). Regulation of Oct transcription activity by O-GlcNAcylation. FASEB J. (in press), doi:10.1096/fj.12-220897 [DOI] [PMC free article] [PubMed]
- Karwacki-Neisius V., Göke J., Osorno R., Halbritter F., Ng J. H., Weiße A. Y., Wong F. C., Gagliardi A., Mullin N. P., Festuccia N., et al. (2013). Reduced oct4 expression directs a robust pluripotent state with distinct signaling activity and increased enhancer occupancy by oct4 and nanog. Cell Stem Cell 12, 531–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehler J., Tolkunova E., Koschorz B., Pesce M., Gentile L., Boiani M., Lomelí H., Nagy A., McLaughlin K. J., Schöler H. R., et al. (2004). Oct4 is required for primordial germ cell survival. EMBO Rep. 5, 1078–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemler I., Schreiber E., Müller M. M., Matthias P., Schaffner W. (1989). Octamer transcription factors bind to two different sequence motifs of the immunoglobulin heavy chain promoter. EMBO J. 8, 2001–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim U., Qin X. F., Gong S., Stevens S., Luo Y., Nussenzweig M., Roeder R. G. (1996). The B-cell-specific transcription coactivator OCA-B/OBF-1/Bob-1 is essential for normal production of immunoglobulin isotypes. Nature 383, 542–547 [DOI] [PubMed] [Google Scholar]
- Kim J. B., Sebastiano V., Wu G., Araúzo-Bravo M. J., Sasse P., Gentile L., Ko K., Ruau D., Ehrich M., van den Boom D., et al. (2009). Oct4-induced pluripotency in adult neural stem cells. Cell 136, 411–419 [DOI] [PubMed] [Google Scholar]
- Klemm J. D., Rould M. A., Aurora R., Herr W., Pabo C. O. (1994). Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell 77, 21–32 [DOI] [PubMed] [Google Scholar]
- Lachnit M., Kur E., Driever W. (2008). Alterations of the cytoskeleton in all three embryonic lineages contribute to the epiboly defect of Pou5f1/Oct4 deficient MZspg zebrafish embryos. Dev. Biol. 315, 1–17 [DOI] [PubMed] [Google Scholar]
- Landolfi N. F., Capra J. D., Tucker P. W. (1986). Interaction of cell-type-specific nuclear proteins with immunoglobulin VH promoter region sequences. Nature 323, 548–551 [DOI] [PubMed] [Google Scholar]
- LaRonde-LeBlanc N. A., Wolberger C. (2003). Structure of HoxA9 and Pbx1 bound to DNA: Hox hexapeptide and DNA recognition anterior to posterior. Genes Dev. 17, 2060–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Weber M., Salgame P., Hu C., Davydov I. V., Laur O., Klevenz S., Krammer P. H. (1998). Th2-specific protein/DNA interactions at the proximal nuclear factor-AT site contribute to the functional activity of the human IL-4 promoter. J. Immunol. 161, 1380–1389 [PubMed] [Google Scholar]
- Liang J., Wan M., Zhang Y., Gu P., Xin H., Jung S. Y., Qin J., Wong J., Cooney A. J., Liu D., et al. (2008). Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat. Cell Biol. 10, 731–739 [DOI] [PubMed] [Google Scholar]
- Lin Y., Yang Y., Li W., Chen Q., Li J., Pan X., Zhou L., Liu C., Chen C., He J., et al. (2012). Reciprocal regulation of Akt and Oct4 promotes the self-renewal and survival of embryonal carcinoma cells. Mol. Cell 48, 627–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox J., Shakya A., South S., Shelton D., Andersen J. N., Chidester S., Kang J., Gligorich K. M., Jones D. A., Spangrude G. J., et al. (2012). Transcription factor Oct1 is a somatic and cancer stem cell determinant. PLoS Genet. 8, e1003048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhas A. N., Lee C. F., Vaux D. J. (2009). Lamin B1 controls oxidative stress responses via Oct-1. J. Cell Biol. 184, 45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui S., Nakatake Y., Toyooka Y., Shimosato D., Yagi R., Takahashi K., Okochi H., Okuda A., Matoba R., Sharov A. A., et al. (2007). Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat. Cell Biol. 9, 625–635 [DOI] [PubMed] [Google Scholar]
- Matsumoto I., Ohmoto M., Narukawa M., Yoshihara Y., Abe K. (2011). Skn-1a (Pou2f3) specifies taste receptor cell lineage. Nat. Neurosci. 14, 685–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna M., Monte P., Helfand S. L., Woodard C., Carlson J. (1989). A simple chemosensory response in Drosophila and the isolation of acj mutants in which it is affected. Proc. Natl. Acad. Sci. USA 86, 8118–8122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A., Mikkelsen T. S., Gu H., Wernig M., Hanna J., Sivachenko A., Zhang X., Bernstein B. E., Nusbaum C., Jaffe D. B., et al. (2008). Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454, 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monuki E. S., Weinmaster G., Kuhn R., Lemke G. (1989). SCIP: a glial POU domain gene regulated by cyclic AMP. Neuron 3, 783–793 [DOI] [PubMed] [Google Scholar]
- Nakai S., Kawano H., Yudate T., Nishi M., Kuno J., Nagata A., Jishage K., Hamada H., Fujii H., Kawamura K., et al. (1995). The POU domain transcription factor Brn-2 is required for the determination of specific neuronal lineages in the hypothalamus of the mouse. Genes Dev. 9, 3109–3121 [DOI] [PubMed] [Google Scholar]
- Nakshatri H., Nakshatri P., Currie R. A. (1995). Interaction of Oct-1 with TFIIB. Implications for a novel response elicited through the proximal octamer site of the lipoprotein lipase promoter. J. Biol. Chem. 270, 19613–19623 [DOI] [PubMed] [Google Scholar]
- Neumann C. J., Cohen S. M. (1998). Boundary formation in Drosophila wing: Notch activity attenuated by the POU protein Nubbin. Science 281, 409–413 [DOI] [PubMed] [Google Scholar]
- Ng M., Diaz-Benjumea F. J., Cohen S. M. (1995). Nubbin encodes a POU-domain protein required for proximal-distal patterning in the Drosophila wing. Development 121, 589–599 [DOI] [PubMed] [Google Scholar]
- Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Schöler H., Smith A. (1998). Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95, 379–391 [DOI] [PubMed] [Google Scholar]
- Nieto L., Joseph G., Stella A., Henri P., Burlet-Schiltz O., Monsarrat B., Clottes E., Erard M. (2007). Differential effects of phosphorylation on DNA binding properties of N Oct-3 are dictated by protein/DNA complex structures. J. Mol. Biol. 370, 687–700 [DOI] [PubMed] [Google Scholar]
- Okamoto K., Okazawa H., Okuda A., Sakai M., Muramatsu M., Hamada H. (1990). A novel octamer binding transcription factor is differentially expressed in mouse embryonic cells. Cell 60, 461–472 [DOI] [PubMed] [Google Scholar]
- Ong C. T., Cheng H. T., Chang L. W., Ohtsuka T., Kageyama R., Stormo G. D., Kopan R. (2006). Target selectivity of vertebrate notch proteins. Collaboration between discrete domains and CSL-binding site architecture determines activation probability. J. Biol. Chem. 281, 5106–5119 [DOI] [PubMed] [Google Scholar]
- Onichtchouk D. (2012). Pou5f1/oct4 in pluripotency control: insights from zebrafish. Genesis 50, 75–85 [DOI] [PubMed] [Google Scholar]
- Pang Z. P., Yang N., Vierbuchen T., Ostermeier A., Fuentes D. R., Yang T. Q., Citri A., Sebastiano V., Marro S., Südhof T. C., et al. (2011). Induction of human neuronal cells by defined transcription factors. Nature 476, 220–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo M., Lang B., Yu L., Prosser H., Bradley A., Babu M. M., Choudhary J. (2010). An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell 6, 382–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearse R. V., 2nd, Drolet D. W., Kalla K. A., Hooshmand F., Bermingham J. R., Jr, Rosenfeld M. G. (1997). Reduced fertility in mice deficient for the POU protein sperm-1. Proc. Natl. Acad. Sci. USA 94, 7555–7560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnusamy M. P., Deb S., Dey P., Chakraborty S., Rachagani S., Senapati S., Batra S. K. (2009). RNA polymerase II associated factor 1/PD2 maintains self-renewal by its interaction with Oct3/4 in mouse embryonic stem cells. Stem Cells 27, 3001–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Préfontaine G. G., Walther R., Giffin W., Lemieux M. E., Pope L., Haché R. J. (1999). Selective binding of steroid hormone receptors to octamer transcription factors determines transcriptional synergism at the mouse mammary tumor virus promoter. J. Biol. Chem. 274, 26713–26719 [DOI] [PubMed] [Google Scholar]
- Reményi A., Tomilin A., Pohl E., Lins K., Philippsen A., Reinbold R., Schöler H. R., Wilmanns M. (2001). Differential dimer activities of the transcription factor Oct-1 by DNA-induced interface swapping. Mol. Cell 8, 569–580 [DOI] [PubMed] [Google Scholar]
- Rodda D. J., Chew J. L., Lim L. H., Loh Y. H., Wang B., Ng H. H., Robson P. (2005). Transcriptional regulation of nanog by OCT4 and SOX2. J. Biol. Chem. 280, 24731–24737 [DOI] [PubMed] [Google Scholar]
- Rosner M. H., Vigano M. A., Ozato K., Timmons P. M., Poirier F., Rigby P. W., Staudt L. M. (1990). A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature 345, 686–692 [DOI] [PubMed] [Google Scholar]
- Scheidereit C., Heguy A., Roeder R. G. (1987). Identification and purification of a human lymphoid-specific octamer-binding protein (OTF-2) that activates transcription of an immunoglobulin promoter in vitro. Cell 51, 783–793 [DOI] [PubMed] [Google Scholar]
- Scheidereit C., Cromlish J. A., Gerster T., Kawakami K., Balmaceda C. G., Currie R. A., Roeder R. G. (1988). A human lymphoid-specific transcription factor that activates immunoglobulin genes is a homoeobox protein. Nature 336, 551–557 [DOI] [PubMed] [Google Scholar]
- Schild-Poulter C., Shih A., Tantin D., Yarymowich N. C., Soubeyrand S., Sharp P. A., Haché R. J. (2007). DNA-PK phosphorylation sites on Oct-1 promote cell survival following DNA damage. Oncogene 26, 3980–3988 [DOI] [PubMed] [Google Scholar]
- Schöler H. R. (1991). Octamania: the POU factors in murine development. Trends Genet. 7, 323–329 [DOI] [PubMed] [Google Scholar]
- Schöler H. R., Ruppert S., Suzuki N., Chowdhury K., Gruss P. (1990). New type of POU domain in germ line-specific protein Oct-4. Nature 344, 435–439 [DOI] [PubMed] [Google Scholar]
- Schonemann M. D., Ryan A. K., McEvilly R. J., O’Connell S. M., Arias C. A., Kalla K. A., Li P., Sawchenko P. E., Rosenfeld M. G. (1995). Development and survival of the endocrine hypothalamus and posterior pituitary gland requires the neuronal POU domain factor Brn-2. Genes Dev. 9, 3122–3135 [DOI] [PubMed] [Google Scholar]
- Schubart D. B., Rolink A., Kosco-Vilbois M. H., Botteri F., Matthias P. (1996). B-cell-specific coactivator OBF-1/OCA-B/Bob1 required for immune response and germinal centre formation. Nature 383, 538–542 [DOI] [PubMed] [Google Scholar]
- Segil N., Roberts S. B., Heintz N. (1991). Mitotic phosphorylation of the Oct-1 homeodomain and regulation of Oct-1 DNA binding activity. Science 254, 1814–1816 [DOI] [PubMed] [Google Scholar]
- Shakya A., Cooksey R., Cox J. E., Wang V., McClain D. A., Tantin D. (2009). Oct1 loss of function induces a coordinate metabolic shift that opposes tumorigenicity. Nat. Cell Biol. 11, 320–327 [DOI] [PubMed] [Google Scholar]
- Shakya A., Kang J., Chumley J., Williams M. A., Tantin D. (2011). Oct1 is a switchable, bipotential stabilizer of repressed and inducible transcriptional states. J. Biol. Chem. 286, 450–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazaki T., Arsenijevic Y., Ryan A. K., Rosenfeld M. G., Weiss S. (1999). A role for the POU-III transcription factor Brn-4 in the regulation of striatal neuron precursor differentiation. EMBO J. 18, 444–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Eckerle S., Onichtchouk D., Marrs J. A., Nitschke R., Driever W. (2013). Pou5f1-dependent EGF expression controls E-cadherin endocytosis, cell adhesion, and zebrafish epiboly movements. Dev. Cell 24, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudt L. M., Singh H., Sen R., Wirth T., Sharp P. A., Baltimore D. (1986). A lymphoid-specific protein binding to the octamer motif of immunoglobulin genes. Nature 323, 640–643 [DOI] [PubMed] [Google Scholar]
- Staudt L. M., Clerc R. G., Singh H., LeBowitz J. H., Sharp P. A., Baltimore D. (1988). Cloning of a lymphoid-specific cDNA encoding a protein binding the regulatory octamer DNA motif. Science 241, 577–580 [DOI] [PubMed] [Google Scholar]
- Ström A. C., Forsberg M., Lillhager P., Westin G. (1996). The transcription factors Sp1 and Oct-1 interact physically to regulate human U2 snRNA gene expression. Nucleic Acids Res. 24, 1981–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm R., Baumruker T., Franza B. R., Jr, Herr W. (1987). A 100-kD HeLa cell octamer binding protein (OBP100) interacts differently with two separate octamer-related sequences within the SV40 enhancer. Genes Dev. 1, 1147–1160 [DOI] [PubMed] [Google Scholar]
- Sturm R. A., Das G., Herr W. (1988). The ubiquitous octamer-binding protein Oct-1 contains a POU domain with a homeo box subdomain. Genes Dev. 2 12A, 1582–1599 [DOI] [PubMed] [Google Scholar]
- Sugitani Y., Nakai S., Minowa O., Nishi M., Jishage K., Kawano H., Mori K., Ogawa M., Noda T. (2002). Brn-1 and Brn-2 share crucial roles in the production and positioning of mouse neocortical neurons. Genes Dev. 16, 1760–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Rohdewohld H., Neuman T., Gruss P., Schöler H. R. (1990). Oct-6: a POU transcription factor expressed in embryonal stem cells and in the developing brain. EMBO J. 9, 3723–3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- Tantin D., Schild-Poulter C., Wang V., Haché R. J., Sharp P. A. (2005). The octamer binding transcription factor Oct-1 is a stress sensor. Cancer Res. 65, 10750–10758 [DOI] [PubMed] [Google Scholar]
- Tantin D., Gemberling M., Callister C., Fairbrother W. G. (2008). High-throughput biochemical analysis of in vivo location data reveals novel distinct classes of POU5F1(Oct4)/DNA complexes. Genome Res. 18, 631–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia N., Reinhardt P., Duemmler A., Wu G., Araúzo-Bravo M. J., Esch D., Greber B., Cojocaru V., Rascon C. A., Tazaki A., et al. (2012). Reprogramming to pluripotency is an ancient trait of vertebrate Oct4 and Pou2 proteins. Nat. Commun. 3, 1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodorou E., Dalembert G., Heffelfinger C., White E., Weissman S., Corcoran L., Snyder M. (2009). A high throughput embryonic stem cell screen identifies Oct-2 as a bifunctional regulator of neuronal differentiation. Genes Dev. 23, 575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichy A. L., Ray A., Carlson J. R. (2008). A new Drosophila POU gene, pdm3, acts in odor receptor expression and axon targeting of olfactory neurons. J. Neurosci. 28, 7121–7129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolkunova E., Malashicheva A., Parfenov V. N., Sustmann C., Grosschedl R., Tomilin A. (2007). PIAS proteins as repressors of Oct4 function. J. Mol. Biol. 374, 1200–1212 [DOI] [PubMed] [Google Scholar]
- van den Berg D. L., Zhang W., Yates A., Engelen E., Takacs K., Bezstarosti K., Demmers J., Chambers I., Poot R. A. (2008). Estrogen-related receptor beta interacts with Oct4 to positively regulate Nanog gene expression. Mol. Cell. Biol. 28, 5986–5995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg D. L., Snoek T., Mullin N. P., Yates A., Bezstarosti K., Demmers J., Chambers I., Poot R. A. (2010). An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell 6, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel D. A., Udalova I. A., De Silva A. P., McGovern D. P., Kinouchi Y., Hull J., Lench N. J., Cardon L. R., Carey A. H., Jewell D. P., et al. (2002). Inflammatory bowel disease is associated with a TNF polymorphism that affects an interaction between the OCT1 and NF(-kappa)B transcription factors. Hum. Mol. Genet. 11, 1281–1289 [DOI] [PubMed] [Google Scholar]
- Van Hoof D., Muñoz J., Braam S. R., Pinkse M. W., Linding R., Heck A. J., Mummery C. L., Krijgsveld J. (2009). Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell 5, 214–226 [DOI] [PubMed] [Google Scholar]
- Verrijzer C. P., van Oosterhout J. A., van der Vliet P. C. (1992). The Oct-1 POU domain mediates interactions between Oct-1 and other POU proteins. Mol. Cell. Biol. 12, 542–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster D. M., Teo C. F., Sun Y., Wloga D., Gay S., Klonowski K. D., Wells L., Dougan S. T. (2009). O-GlcNAc modifications regulate cell survival and epiboly during zebrafish development. BMC Dev. Biol. 9, 28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F., Schöler H. R., Atchison M. L. (2007). Sumoylation of Oct4 enhances its stability, DNA binding, and transactivation. J. Biol. Chem. 282, 21551–21560 [DOI] [PubMed] [Google Scholar]
- Wei Z., Yang Y., Zhang P., Andrianakos R., Hasegawa K., Lyu J., Chen X., Bai G., Liu C., Pera M., et al. (2009). Klf4 interacts directly with Oct4 and Sox2 to promote reprogramming. Stem Cells 27, 2969–2978 [DOI] [PubMed] [Google Scholar]
- Xu H. M., Liao B., Zhang Q. J., Wang B. B., Li H., Zhong X. M., Sheng H. Z., Zhao Y. X., Zhao Y. M., Jin Y. (2004). Wwp2, an E3 ubiquitin ligase that targets transcription factor Oct-4 for ubiquitination. J. Biol. Chem. 279, 23495–23503 [DOI] [PubMed] [Google Scholar]
- Yang X., Yeo S., Dick T., Chia W. (1993). The role of a Drosophila POU homeo domain gene in the specification of neural precursor cell identity in the developing embryonic central nervous system. Genes Dev. 7, 504–516 [DOI] [PubMed] [Google Scholar]
- Yeap L. S., Hayashi K., Surani M. A. (2009). ERG-associated protein with SET domain (ESET)-Oct4 interaction regulates pluripotency and represses the trophectoderm lineage. Epigenetics Chromatin 2, 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P., Han J., Guo G., Orlov Y. L., Huss M., Loh Y. H., Yaw L. P., Robson P., Lim B., Ng H. H. (2009). Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev. 23, 2507–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Liao B., Xu M., Jin Y. (2007). Post-translational modification of POU domain transcription factor Oct-4 by SUMO-1. FASEB J. 21, 3042–3051 [DOI] [PubMed] [Google Scholar]
- Zheng L., Roeder R. G., Luo Y. (2003). S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell 114, 255–266 [DOI] [PubMed] [Google Scholar]
- Zhu S., Li W., Zhou H., Wei W., Ambasudhan R., Lin T., Kim J., Zhang K., Ding S. (2010). Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell 7, 651–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwilling S., Dieckmann A., Pfisterer P., Angel P., Wirth T. (1997). Inducible expression and phosphorylation of coactivator BOB.1/OBF.1 in T cells. Science 277, 221–225 [DOI] [PubMed] [Google Scholar]