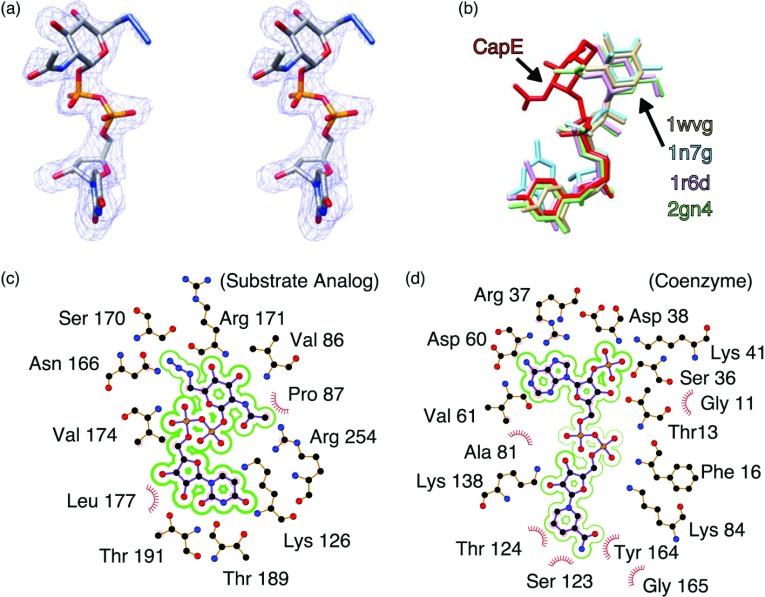
Figure 3. Conformation of substrate analogue and coenzyme.
(a) Stereoview of the substrate analogue UDP-6N3-GlcNAc. The electron density correspond the sigma-A weighted 2Fo–Fc map contoured at 1.0 σ. The substrate analogue is depicted with CPK colours. (b) Conformation of various nucleotide-sugars bound to representative SDR enzymes. The panel depicts CapE (this study, red); CDP-β-D-xylose bound to CDP-D-glucose 4,6-dehydratase (1wvg, yellow); GDP-rhamnose bound to GDP-mannose 4,6-dehydratase (1n7g, cyan); dTDP-D-glucose bound to dTDP-glucose 4,6-dehydratase (D128N/E129Q mutein, 1r6d, pink); and UDP-GlcNAc bound to FlaA1 (UDP-GlcNAc inverting 4,6-dehydratase, 2gn4, green). (c) Residue environment around the substrate analogue in wild-type CapE and (d) in the binding pocket of the coenzyme in K126E. Panels (c) and (d) were generated with the program LIGPLOT [51].