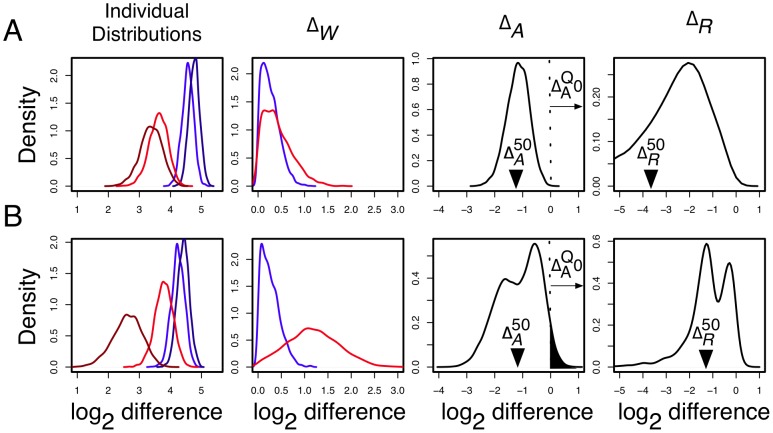
Figure 3. Approximate ANOVA via Absolute and Relative fold differences.
The figure shows how the method explicitly accounts for the within-condition dispersion using as an example two genes with similar absolute fold differences (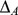 ) of -1.17 and -1.13 but very different relative fold differences (
) of -1.17 and -1.13 but very different relative fold differences (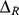 ) and
) and 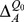 values in the L-K dataset. Dirichlet sampled distributions are generated from the raw read counts as described in the text. These distributions are log-transformed and the noninformative subspace is removed. Posterior distributions of
values in the L-K dataset. Dirichlet sampled distributions are generated from the raw read counts as described in the text. These distributions are log-transformed and the noninformative subspace is removed. Posterior distributions of 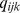 are shown for
are shown for 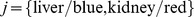 and for replicates
and for replicates 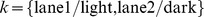 . Both genes are abundantly expressed in this dataset, with median expression levels between
. Both genes are abundantly expressed in this dataset, with median expression levels between 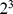 and
and 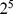 greater than the mean across-gene expression level.
greater than the mean across-gene expression level. 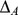 is computed by randomly sampling one of the red distributions, randomly sampling one of the blue distributions, and subtracting for all pairs of between-condition distributions.
is computed by randomly sampling one of the red distributions, randomly sampling one of the blue distributions, and subtracting for all pairs of between-condition distributions. 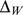 is computed by sampling a light and a dark from each red and blue distributions, subtracting light and dark, and selecting the difference with the greatest magnitude.
is computed by sampling a light and a dark from each red and blue distributions, subtracting light and dark, and selecting the difference with the greatest magnitude. 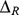 is computed as the ratio of a single realization of
is computed as the ratio of a single realization of 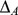 and
and 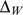 , and is computed for each realization of
, and is computed for each realization of 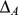 and
and 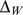 . The
. The 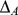 distribution is narrower in A than in B implying a greater precision in estimating this value. This precision can be estimated by
distribution is narrower in A than in B implying a greater precision in estimating this value. This precision can be estimated by 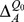 , the quantile of zero in
, the quantile of zero in 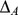 , which is shown graphically as the black-filled area under the
, which is shown graphically as the black-filled area under the 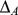 distribution curves. The
distribution curves. The 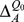 values are 0.0001 and 0.035 in panels A and B respectively. The vertical arrows show the median values of
values are 0.0001 and 0.035 in panels A and B respectively. The vertical arrows show the median values of 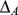 and
and 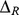 . Thus, the between-condition expression values for the gene in Panel A are scored as separable by ALDEx (
. Thus, the between-condition expression values for the gene in Panel A are scored as separable by ALDEx (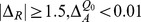 ) but not for the gene in Panel B (
) but not for the gene in Panel B (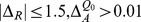 ). These conclusions agree with inspection-based intuition from examining the initial adjusted log-expression distributions that are shown in the left panel.
). These conclusions agree with inspection-based intuition from examining the initial adjusted log-expression distributions that are shown in the left panel.