Abstract
For decades, immunoassays have provided the framework for protein biomarker studies in clinical medicine and in therapeutic monitoring for drug development. At the same time, investigators have uncovered many issues that make immunoassays unreliable in many human serum and plasma samples. LC-MS/MS after tryptic digestion of proteins is potentially an attractive solution, but the sensitivity of the method is not sufficient to measure many important low-abundance proteins directly. The use of antipeptide antibodies to immunoenrich peptides of interest can improve the sensitivity of the approach, greatly simplify the matrix enabling shortened chromatographic runs, and facilitate the multiplexed quantification of analytes, which could reduce the costs of quantitative protein measurements in complex specimens. We provide an overview of the method and the steps needed to develop an assay. In addition, we review the efforts to make this method generally more applicable.
Immunoassays (IAs) have been used in research, industry and clinical settings for decades. While they have shown great utility, they also have known flaws [1]. Chief among these flaws are poor concordance between assays and interferences [2–4]. The poor concordance between different manufacturers stems from the different proprietary antibodies used to detect different epitopes in each assay, and the influence of natural biological diversity, such as post-translational modifications and single nucleotide polymorphisms, and variable cross-reactivities of these different immunoassay reagent antibodies to off-target proteins. Additionally, a variety of interferences have been observed, including antireagent antibodies and endogenous autoantibodies, which can generate incorrect results in an IA and have caused patients harm [5,6]. The indirect, nonspecific signals measured in immunoassays thus mask the true identity of the molecule being detected, whether it is a target analyte, off-target protein or some other interference.
Considering these faults, a viable alternative to IAs may be tryptic digestion of proteins to peptides, followed by analyte-specific peptide detection and quantification by LC-MS/MS. While this method can easily detect abundant proteins in a quantitative manner, it is not nearly sensitive enough for low-abundance proteins [7–18]. As a result, enrichment of target analytes may be useful in the quantification of low-abundance proteins. One method of enrichment and quantification has been termed stable-isotope standards and capture by antipeptide antibodies (SISCAPA), which uses immunoaffinity enrichment of target peptides and labeled internal standard peptides in the quantification of proteins in proteolytically digested complex mixtures [19]. In contrast to IAs, this technique is not subject to interference by endogenous immunoglobulins, because those antibodies are destroyed by tryptic digestion. There is now strong precedence established for using peptides as surrogates for accurate protein quantification [7,11,18] and there are ample data demonstrating that SISCAPA is capable of achieving LOD similar to IAs [20–23]. There are additional benefits of SISCAPA LC-MS/MS assays, namely the specificity of the system and the ability to multiplex analytes. By directly detecting peptide analyte, SISCAPA assays avoid many of the pitfalls of immunoassays that manufacturers spend a great deal of money trying to avoid. In this review we describe how SISCAPA assays work, describe a potential workflow for assay development, discuss potential pitfalls and consider the future of this technology.
Overview of the SISCAPA workflow
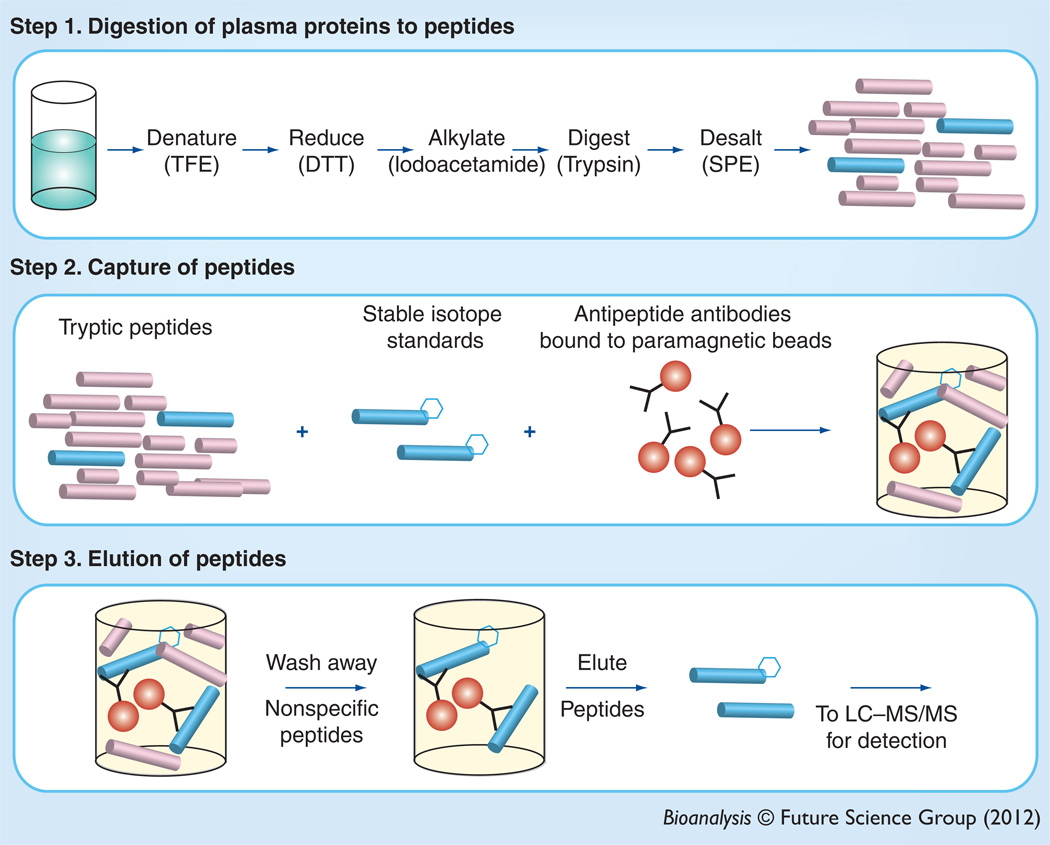
SISCAPA is a multistep process that can be effectively used to enrich and quantify low-abundance peptides from complex matrices such as plasma (Figure 1). Importantly, peptides from higher abundance proteins can also be enriched and quantified in similar assays [24]. The process begins with tryptic digestion of a sample, which cleaves proteins into peptides. This step is a well-known source of variability in the process [25,26]. The digested sample is incubated with antipeptide antibodies bound to a solid phase, typically paramagnetic beads. The internal standard (IS), which is a stable isotope-labeled peptide, is included during this incubation. Peptides liberated during the digestion of endogenous target analyte and the corresponding IS peptides are bound to the solid phase, and the unbound matrix is washed away. This washing step significantly decreases the complexity of the sample that will be subsequently introduced to the MS system, thus greatly increasing the chances that the peptides of interest will be detectable.
Figure 1. Peptide immunoaffinity enrichment after trypsin digestion.
Peptides are liberated from proteins in complex samples after denaturation, reduction and alkylation. Tryptic peptides are then enriched using antipeptide antibodies bound to a paramagnetic solid-phase support.
DTT: Dithiothreitol; TFE: Trifluoroethanol.
Overview of LC–MS/MS
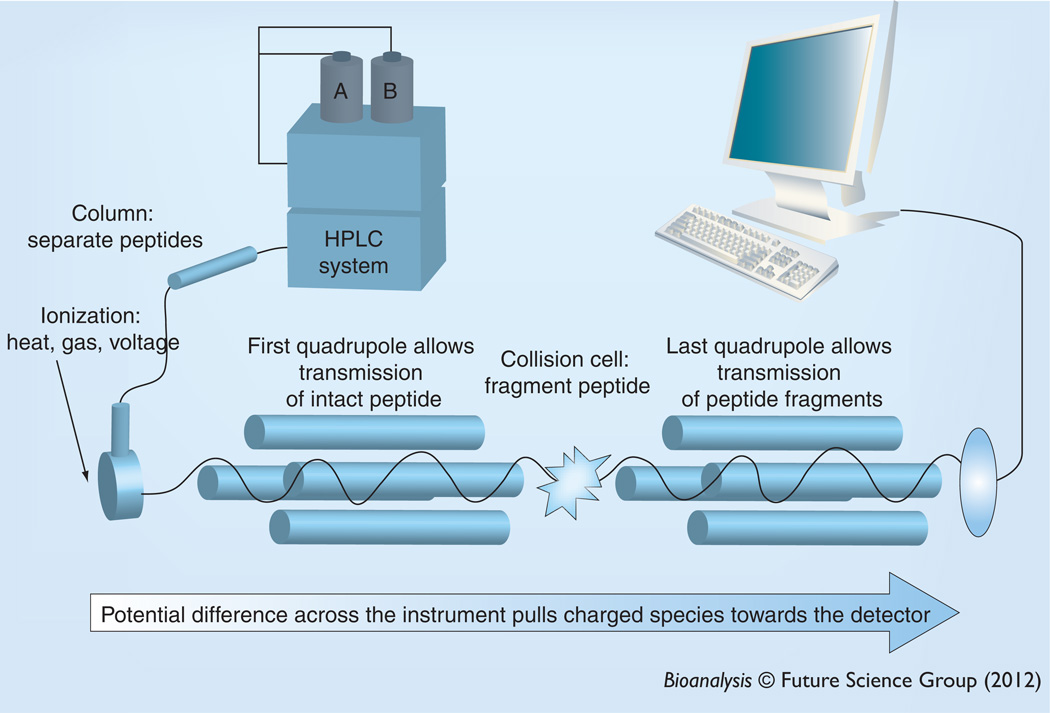
Peptides are detected and quantified using MS (Figure 2). Peptides are first separated from one another and other matrix components using HPLC, most commonly using reverse-phase chemistry. Eluted analytes are ionized at the instrument source using ESI. During this process, heat, an inert gas and voltage are applied to eluent, which results in charged analyte ions. To quantify the peptide of interest in a triple quadrupole mass spectrometer, the intact peptide is selected in the first mass filter, the peptide is fragmented using inert gas in the collision cell, and specific fragments are selected in the collision cell. Ions are pulled into and through the MS system to the detector by a voltage gradient. In the first quadrupole, RF voltages are used to filter and select for the m/zof the intact analyte of interest. All other mlz will not pass through the quadrupole. The selected ions with proper mlz exit the first quadrupole and enter the collision cell where they collide with controlled amounts of inert gas under the influence of specific optimized voltages. This dissociates the analyte into fragments, which if charged will enter the next quadrupole. Similar to the first quadrupole, this also serves as a mass filter and is set to transmit fragment ions with specific m/z that will pass through and hit the detector. The detector then sends a signal to the computer to say that the proper precursor m/z–fragment m/z pair was present and can provide a direct measure of the number of ions that have passed through the two mass filters. This pair of m/z for the first and last quadrupoles is called a transition and a MS can cycle through several transitions in a mode called multiple reaction monitoring (MRM). In this review we only address the use of a MS/MS system equipped with an ESI or nanospray ioniziation interface, but it is not the only available option. Instruments with MALDI sources and with orbitrap, TOF or TOF–TOF analyzers may also have utility for SISCAPA assays [4,27–29].
Figure 2. LC-MS/MS.
Peptides are separated using HPLC and analyzed using two mass analyzers. In the first quadrupole mass analyzer, the intact peptide is selected and then fragmented using collision-induced dissociation in the collision cell and fragments are selected in the last quadrupole. Any ions that strike the detector have the correct m/z of the intact precursor and the correct fragment m/z.
Components
To perform SISCAPA LC–MS/MS methods, various components are required and include peptides, solid-phase supports for affinity enrichment, antibodies, standard materials and IS, each of which must be identified, produced and validated. In this section, we briefly discuss a process by which each of these components can be identified and produced.
Peptide selection
Empiric peptide selection
The first step in the development of a quantitative SISCAPA assay is the selection of peptides that serve as the surrogates for protein analytes that can be accomplished empirically. The ideal peptides are those that ionize well in the MS system and do not have homologues elsewhere in the genome. Peptides that fit in this category are often called proteotypic peptides. To select proteotypic peptides that are reliably liberated during proteolytic digestion empirically, a source of the protein to be quantified needs to be identified. For example, the protein of interest may already be a component of an IA, which is to be replaced, may be available commercially, may be purified it from its natural source, or may be expressed recombinantly. The protein is digested and, ideally, analyzed using the LC–MS/MS system that will be used for the final assay. Using the same instrument that will be used for quantification helps identify peptides that will be robustly ionized, fragmented and detected in the final assay, which may vary on different MS platforms. We have found that a very efficient workflow to choose peptides starts with Skyline, a free, open-source software product developed in the MacCoss laboratory [30,31]. In this workflow, a protein sequence file (e.g., from the NCBI database) is imported into Skyline and digested in silico and used to generate a list of tryptic peptides from the protein of interest. The software also produces a list of putative MRM transition pairs with theoretically optimized instrument parameters. It is possible to further optimize Skyline software output by selecting ions and charge states that are more typically seen by the instrument in use. The list of MRM transition pairs and instrument parameters can be directly loaded into the instrument software and used to scout for the best peptides. To empirically determine the most robust peptides for quantification, multiple injections of protein digest are made and the precision of peak areas used in the selection process [32].
Ideally, peptides that contain amino acid residues that can be oxidized or deamidated (e.g., methionine, cysteine and tryptophan) should be avoided since this can complicate quantification later. Further screening is required to verify that the potential peptides are unique in the genome and free from post-translational modifications or common coding single nucleotide polymorphisms, as these can lead to inaccurate assay results. This is accomplished by database searching, which has been reviewed by Lange et al., and their excellent review of this process also provides links to the database resources [33]. In the end, a small collection of proteotypic peptides (e.g., three to five peptides) for each analyte is generally chosen, rather than a single peptide, because it is not always possible to know about and therefore avoid every post-translational modification or single nucleotide polymorphism. In addition, monitoring multiple peptides generates a more specific assay and helps prevent unrecognized chromatographic interference in the final assay. Lastly, not all peptides consistently generate a robust immune response [34]. Thus, having more peptides increases the likelihood that antibody production will be successful and the method will be more robust.
Another approach to peptide selection
If purified protein is not available then there are a few other approaches to peptide selection that have been described. For example, analytes may be detected directly from digested samples [7,35]. In this example, a small number of samples expected to contain the analyte of interest are processed and pooled. Injections are made onto the LC–MS/MS system to pick the best peptides using the Skyline workflow. Other approaches use MS/MS and database searching for peptide identification [36]. Alternatively, sample preparation methods such as 2D gel electrophoresis or capture and immunoprecipitation of whole protein by antibodies that have been purchased or pulled from an IA may also help provide material for digest and peptide selection [17]. These alternative workflows can be labor or computationally intensive as compared with digestion of purified analyte because, in a complex sample, there are more interferences that compete for detection by the MS system. There can also be uncertainty about correct peptide identification. Fortunately, there have been important advances in reducing the computational time needed to perform database searches of MS/MS spectra [37,38].
In silico selection of peptides
Another alternative to empirically selecting well-liberated, well-ionized peptides from digested intact proteins is to select proteotypic peptides purely in silico. Databases of peptides and transitions observed in proteomic experiments have been assembled and are easily searchable [39,40]. In addition, Skyline can be used identify putatively useful peptides from proteins. After using these tools, potentially useful peptides can be synthesized and their performance can be evaluated.
Antibodies
Antipeptide antibodies targeting analyte peptides are necessary for the immuno-affinity enrichment of those peptides prior to LC–MS/MS analysis. As a result, this reagent is key to the sensitivity of SISCAPA assays. Several reports have determined the effectiveness of immunoaffinity enrichment of peptides using this approach, which range from about 120-fold [19] to 100–1000-fold [34], and up to 18,000-fold [21,24,41]. Of course, extensive enrichment is most important in detecting and quantifying low-abundance peptides. It is less critical and more difficult to enrich peptides that are already abundant in a digest.
Immunogen
Peptides by themselves are generally too small to be presented as antigens in mammals. To generate an immune response, peptides can be attached as haptens to a carrier protein to make an immunogen. An example carrier protein is keyhole limpet hemocyanin (KLH), which actually serves as both a carrier protein and an adjuvant (i.e., a molecule that helps to activate the immune system). The conjugation of peptide to carrier can be accomplished by gluteraldehyde, which crosslinks amine groups. Alternatively, a cystine linker can be added to the peptide when it is synthesized, and sub-sequently can be bound to maleimide activated KLH. This peptide–KLH mixture is used as the immunogen for generating antibodies in a host system.
Antibody production
Antibodies can be produced in polyclonal or monoclonal systems. The most obvious benefits of monoclonal antibodies as reagents for quantitative assays are that the hybridoma represents a renewable resource and that some properly selected monoclonal antibodies can have higher affinity for their target peptides than analogous polyclonal antibodies. There are other benefits, in that they do not require an additional affinity purification step prior to their use in a SISCAPA assay. However, the development of monoclonal antibodies is expensive in terms of labor (i.e., in screening and selecting a hybridoma) and time (i.e., it takes many months to generate a useful, stable hybridoma). If possible, clones should be screened using a SISCAPA assay, because clones selected by traditional ELISA are not always useful in SISCAPA assays [41]. To save time and money, it is also possible to rely on polyclonal antibodies. The success rate is very high for generating polyclonal antibodies that are useful in SISCAPA assays [42], but, as mentioned above, in some circumstances polyclonal antibodies may not have an affinity strong enough to adequately enrich target peptides and the yield may be small enough to limit their prolonged use in a high-throughput setting. Rabbits are attractive mammals to immunize because they produce antibodies of a higher average affinity than rodents, can produce a relatively large supply of antiserum and because spleens from animals that mount a good immune response can be used to make monoclonal antibodies [43].
Affinity purification & passenger peptide
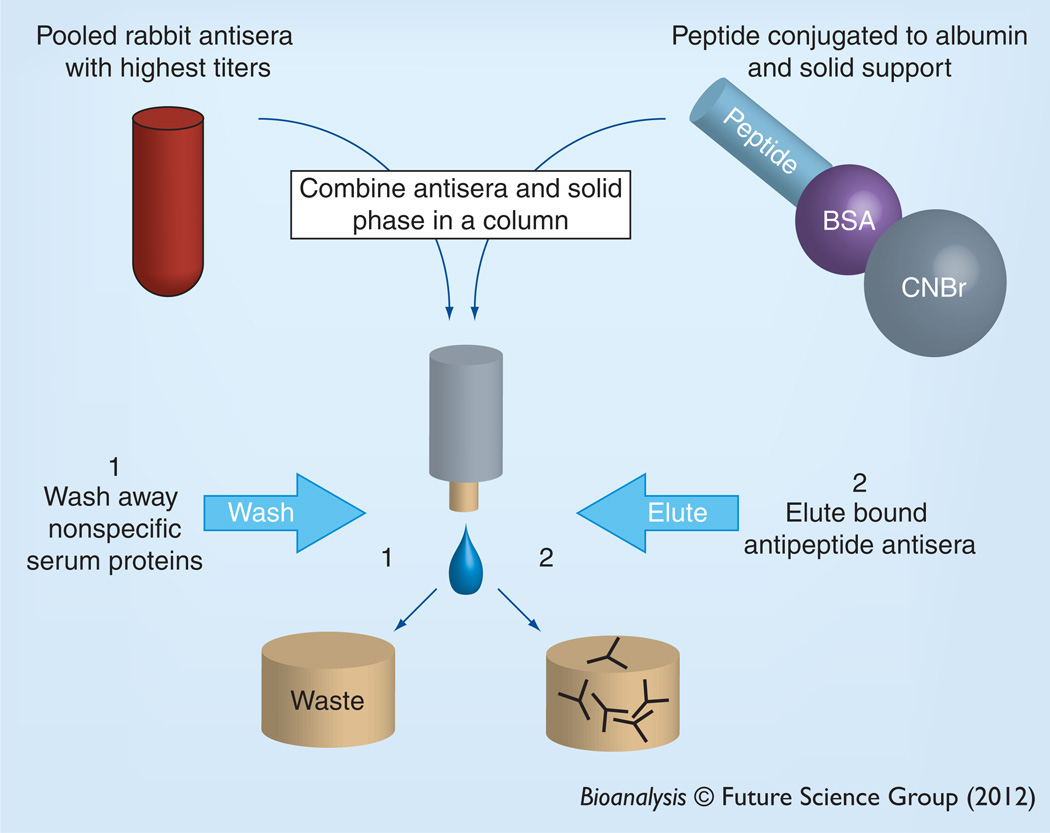
Prior to use in SISCAPA, polyclonal antibodies are affinity purified (Figure 3). The most useful bleeds can be quickly selected by ELISA and pooled. Reagent antibodies can then be purified from this pool using an affinity reagent that can be made by conjugating peptide to albumin, which serves as a carrier protein, and by binding the conjugate to a solid-phase resin such as cyanogen bromide-activated Sepharose® resin. The antiserum pool can then be applied to the Sepharose-albumin-peptide resin and the antipeptide antibodies allowed to bind. Then nonspecific immunoglobulins and other serum components are washed away and the antibody pool, specific to the antigen, is eluted. Importantly, during affinity purification of reagent polyclonal antibody, peptide bound to the solid phase that was used to purify reagent antibodies can slowly dissociate from the solid phase and occupy the antigen binding site of the immunoaffinity-purified reagent antibodies. While this does not cause a problem for most applications, such as western blotting or immunohistochemistry, sensitive MS systems can easily detect the peptide that is copurified. The ‘passenger’ peptide that is co-purified with the antibodies can significantly affect the sensitivity of SISCAPA assays [20] and highlights another potential benefit of monoclonal reagents.
Figure 3. Affinity purification of antipeptide antibodies.
Peptides are conjugated to an albumin carrier protein and linked to a solid-phase support (e.g., Sepharose®). Immunized rabbits are bled and the sera with highest titer are pooled and mixed with the peptides on the solid-phase support. After washing, the bound antibodies are eluted and used in the stable-isotope standards and capture by antipeptide antibodies experiment.
BSA: Bovine serum albumin; CNBr: Cyanogen bromide.
Solid phase
The antibodies used to capture the peptides liberated by trypsin digestion are attached to a solid phase to permit washing. After washing, the peptide may be eluted from the solid phase and analyzed by LC–MS/MS. In most applications published to date, the solid phase used has been paramagnetic beads, and an example of an inline bead-washing apparatus has been described [24]. However, inline methods have also been developed using POROS® resin [19,22]. For paramagnetic beads, two approaches have been deployed:
-
▪
A two-step assay in which the antipeptide antibody is mixed with the tryptic digest and then protein G-coated beads are used to pull peptide–antibody complexes out of the digest [34,41,44];
-
▪
A one-step assay in which antipeptide antibody covalently bound to the solid phase is incubated with the digest to purify target peptides [20,21,45].
In this second type of assay, protein G beads can be used to bind the Fc portion of the antibodies, which holds the antibodies in the proper orientation with the antigen binding site directed away from the bead surface. Then the antibodies can be crosslinked to the beads using commercially available crosslinking reagents. Alternatively, surface-activated beads can be used to covalently bind antipeptide antibodies. The decision to use paramagnetic or polymer beads and the choice of chemistry to use to bind the antipeptide antibody to the beads is largely driven by the final assay format workflow and empirical observations.
IS & calibrators
Stable isotope-labeled IS peptides have the same primary amino sequences as the endogenous peptides selected for analysis. The labeled amino acid residues that are incorporated can have deuterium, 15N or 13C labels that increase the mass of the peptide without affecting the chemical properties. Generally, 15N and 13C affect the chromatographic retention time of the peptide much less than deuterium. Subtle differences in fragmentation patterns are seen with any of the atomic substitutions. The mass shift due to the addition of the isotope labels must be sufficient to allow the MS/MS to distinguish IS from endogenous peptide signal. For example, if monitoring a peptide with a charge state z = +3, an increase of 6 amu would be needed to completely distinguish an isotope-labeled peptide from its endogenous analog. IS peptides are spiked at about a 1:1 ratio with the midpoint of the standard curve, which improves precision and is standard for isotope-dilution quantification by MS. IS peptides spiked after digestion can help control for run-to-run variability due to antibodies, beads, chromatography and MS/MS behaviors. In comparison with labeled IS peptides spiked after digestion and before enrichment, it is easy to imagine that stable isotope-labeled proteins spiked before digestion would be superior in controlling for the complete analysis workflow (i.e., including the digestion step), we could find no examples of this approach in a SISCAPA-type application. Calibrators can be made from either peptides or proteins. Proteins would be generally preferred because their digestion and behavior in the sample matrix will more accurately reflect a real sample. However, calibration curves made from purified proteins spiked into an appropriate matrix may not be ideal and native human serum calibrators could be considered [7].
An example SISCAPA workflow
Day 1
Denaturation, reduction &alkylation
A 20 µl volume of plasma is diluted to 70 µl with 100 mM ammonium bicarbonate. The sample is vortexed and 70 µl of trifluroethanol is added. The sample is vortexed again and incubated for 1 h at 65°C shaking in a heatblock. Cystines are reduced with 5 mM dithiothreitol for 1 h mixing at 65°C. After cooling to room temperature, iodoacetamide is added to 15 mM, quickly vortexed and incubated in the dark for 15–30 min. After alkylation the excess iodoacetamide is scavenged by addition of dithiothreitol to a final concentration of 10 mM. The sample is diluted to 1.4 ml with 100 mM ammonium bicarbonate (for full trypsin activity, trifluroethanol must be diluted to <5%).
Protein digestion
Trypsin at 1:20 protease :protein (w/w) is added twice. The first incubation is 2–4 h and the second is overnight. Both steps are mixing at 37°C.
Preparation of antipeptide beads
A 10 µg volume of antipeptide antibodies are incubated with 20 µl (0.6 mg) of surface-activated beads (e.g., tosyl-activated paramagnetic beads from Invitrogen) and incubated overnight at 37°C mixing.
Day 2
Preparation of antipeptide beads
Beads are blocked in 0.1% bovine serum albumin (w/v) in 200 mM Tris pH 8.5 for 4 h at 37°C, and then washed in 0.1% bovine serum albumin (w/v) phosphate-buffered saline.
Preparation of digests & immunopurification of peptides
Trypsin digestion is stopped with acidification (e.g., to 0.1% formic acid). Peptides are purified using SPE and dried in a centrifugal evaporator. The residue is reconstituted in 0.5 ml phosphate-buffered saline with mixing for 2 h, stable isotope-labeled IS peptide is added and the mixture is transferred to tubes containing antipeptide-coated beads. Samples spiked with IS and antibody-coated beads are incubated with end-over-end agitation overnight at 4°C.
Day 3
Washing beads & eluting peptide
Beads are washed twice with 100 mM ammonium acetate and twice with water. To elute peptides, beads are resuspended in 15 µl 5% acetic acid and incubated for 1–2 h with gentle agitation. The supernatant is then injected on the LC–MS/MS system.
Validation & quality control
Analytical validation of the method includes experiments that define the precision of the assay (within-run and between-run), the LOD and LOQ the linearity of the assay, and evaluation for potential interferences. In proteomics methods, it would be reasonable to start with an evaluation of anticoagulants, lipids (e.g., triglycerides and cholesterol), elevated total protein (e.g., monoclonal gammopathy) and hemolysis. The method should be characterized for ion suppression [46]. Quality control materials should be included in each batch and should evaluate adequacy and consistency of the digestion step as well as instrument performance. Quality control parameters for each sample should include peak area of IS (to evaluate for ion suppression), retention times of analytes (to evaluate HPLC), fragment ratios for each analyte peptide (the ratio of the peak areas of two fragments from the same peptide should be constant across samples, if it is not then there is potential interference with an isobaric interference [32]) and peptide ratios for each protein (the ratio of responses between two peptides from the same protein should be constant across samples, if it is not then there may post-translational modifications or single nucleotide polymorphisms that explain the differences). It is reasonable to compare the SISCAPA LC–MS/ MS method to an alternative IA. It is unusual for such a method comparison to show equivalence, but proportional bias can be less than 50% with a correlation (r2) approximately 0.8 [20,21].
Limitations, troubleshooting & potential pitfalls
Tryptic digestion is variable
Tryptic digestion of complex specimens is variable and typically does not result in complete digestion of all proteins in the sample. This has a significant impact upon the calculation of sample concentrations based on standard curves, particularly the standard curves made from peptides. Previous studies have shown that for some proteins, calibration materials consisting of purified proteins spiked into an appropriate matrix may give inconsistent results day-to-day. A single point calibrator can reduce the impacts of day-to-day variability upon datasets collected over many days [7,47]. Another promising technique for controlling the variability of trypsin digestion is to include an IS protein as part of signal normalization and calibration [18,48,49].
LOD/sensitivity
Even with immunoaffinity peptide purification after tryptic digest, detecting analytes of low femtomolar (ng/ml) concentrations from actual samples with complex matrices can present a challenge. Many previous studies using SISCAPA have determined the LOD of the assay by using peptides spiked into samples after digestion, which are then enriched by immunoaffinity and detected by LC–MS/MS. As mentioned above, because tryptic digestion does not generally go to completion, the amount of peptide available for immunoaffinity enrichment in an actual analysis will be lower than in the peptide-spiking experiment. To improve the LOD of an assay, it is possible to scale up digestion volume, but this becomes impractical at a certain volume of starting material. Additionally, increasing the amount of initial sample does not proportionally increase the amount of peptide liberated during digestion [34]. Given the LOD needed for clinical use and for research settings, we need more sensitive MS systems if we are to be successful. Various promising technologies are being developed [50].
Depletion of abundant proteins
To simplify the complex matrix of serum or plasma prior to proteolytic digestion, it is possible to deplete samples of high abundance proteins. There are pros and cons to the use of depletion [51], but theoretically the sample matrix is simplified and the chances of detecting the analyte by LC–MS/MS are improved. Of course, albumin-bound or immunoglobulin-bound proteins would be at least partially depleted in the process, which would eliminate any benefit of simplifying the matrix. In addition, sample depletion columns are expensive and may not be reusable.
Immunopurification of intact proteins
The enrichment of target analyte can also be performed from undigested sample. One of the most useful approaches uses antibodies. There are many examples of this approach in the literature, some that measure intact proteins, which is limited to proteins of small molecular weight [52–57], and others that use trypsin digestion and quantification of peptides as a surrogate for protein quantification [58–60]. Similar to SISCAPA, harnessing the power of antibodies allows direct detection of analyte of low-abundance proteins using MS, but it must be remembered that abundant binding proteins and autoantibodies to endogenous proteins could block the epitopes targeted by the antibodies. Regardless, immuno-MS assays will likely be important.
Future perspective
Over the next 5-10 years, SISCAPA LC–MS/MS methods will replace certain problematic I As. At the same time, SISCAPA LC–MS/MS assays will be developed to measure and clinically validate novel biomarkers. To position the field to make this change, vendors are making platforms to help automate sample preparation, developing more sensitive instruments and the NIH are spending money to develop the antibody reagents needed for the assays [101].
Automation should allow for more precise assays, ideally with better between-site variability [25,26], less sample handling, higher throughput and shortened method development times [34,41,44]. The ability of automated pipetting stations to prepare reagents in a highly multiplexed fashion would make it easier to assemble panels of assays for each sample.
Alternative reagents to antibodies include aptamers, which are nucleic acid molecules that can be used as high-affinity capture reagents before LC–MS/MS [61]. New instrumentation that improves sensitivity and increase sample throughput will help bring SISCAPA-LC–MS/MS methods into widespread use. Multichannel inlet systems could improve sample throughput [62].
The groundwork has been laid and the development of the tools necessary to make the SISCAPA LC–MS/MS approach more routine is now needed. This powerful technique will find application in basic research, clinical research, pharmaceutical applications, and the clinical care of patients in need of better assays.
Executive summary.
Background
-
▪
Immunoaffinity peptide enrichment after tryptic digestion can improve the LOD of traditional targeted proteomics assays on LC–MS/MS platforms.
Components
-
▪
Methods for calibration and quality control are still being developed and optimized.
Limitations
-
▪
LOD are currently limited by instrument sensitivity.
Future perspective
-
▪
Automation, better instrumentation and reagent development will allow more complete introduction of the methodology.
Acknowledgments
The author’s laboratory has received research support from Waters and Bruker Daltonics. The writing of this manuscript was partially supported by the NIH (grant P30DK035816).
Key Terms
- Antireagent antibodies
One example of a group of interferences that can cause the association of capture and reporter antibodies in a sandwich immunoassay and lead to falsely elevated signals
- Endogenous autoantibodies
An immunoglobulin produced in vivo that recognizes a protein within the human body that can mask the epitopes on an analyte recognized in an immunoassay
- Tryptic digestion
Proteolytic cleavage using the enzyme trypsin, which cleaves proteins at lysine and arginine residues
- LC-MS/MS
A method in which molecules are separated in the liquid phase and then ionized and analyzed in the gas phase. The mlz of the intact molecules is measured, as are fragments of the molecule after it has been fragmented within the MS system
- SISCAPA
An acronym that refers to the enrichment of an analyte-specific peptide and stable isotope-labeled peptide analog from a tryptic digest using peptide-specific antibodies. Analyte-specific peptides and labeled internal standards are then quantified using LC-MS/MS
- m/z
MS/MS depends on ion optics and only charged molecules are detectable. The physical methods used to separate ions from one another in the MS system rely on differences of mlz
- Passenger peptide
Analyte peptide that is co-purified during the affinity purification of reagent polyclonal antibodies
- Isobaric interference
A molecule that co-elutes with the analyte of interest from the HPLC, which has the same m/z as the intact analyte of interest and has a fragment with the same m/z as one of the analyte fragments
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Hoofnagle AN, Wener MH. The fundamental flaws of immunoassays and potential solutions using tandem mass spectrometry. J. Immunol. Methods. 2009;347(1–2):3–11. doi: 10.1016/j.jim.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rawlins ML, Roberts WL. Performance characteristics of six third-generation assays for thyroid-stimulating hormone. Clin. Chem. 2004;50(12):2338–2344. doi: 10.1373/clinchem.2004.039156. [DOI] [PubMed] [Google Scholar]
- 3.La’ulu SL, Roberts WL. Performance characteristics of five automated CA 19–9 assays. Am. J. Clin. Pathol. 2007;127(3):436–440. doi: 10.1309/H52VET3M6P7GYWG1. [DOI] [PubMed] [Google Scholar]
- 4.Mongia SK, Rawlins ML, Owen WE, Roberts WL. Performance characteristics of seven automated CA 125 assays. Am. J. Clin. Pathol. 2006;125(6):921–927. doi: 10.1309/NBA3-12W0-LANR-XYH9. [DOI] [PubMed] [Google Scholar]
- 5.Rotmensch S, Cole LA. False diagnosis and needless therapy of presumed malignant disease in women with false-positive human chorionic gonadotropin concentrations. Lancet. 2000;355(9205):712–715. doi: 10.1016/S0140-6736(00)01324-6. [DOI] [PubMed] [Google Scholar]
- 6.Morgan BR, Tarter TH. Serum heterophile antibodies interfere with prostate specific antigen test and result in over treatment in a patient with prostate cancer. J. Urology. 2001;166(6):2311–2312. [PubMed] [Google Scholar]
- 7. Agger SA, Marney LC, Hoofnagle AN. Simultaneous quantification of apolipoprotein A-I and apolipoprotein B by liquid-chromatography-multiple reaction-monitoring mass spectrometry. Clin. Chem. 2010;56(12):1804–1813. doi: 10.1373/clinchem.2010.152264.▪ Describes the difficulties associated with calibration of MS methods to measure proteins.
- 8.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell. Proteomics. 2006;5(4):573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Barr JR, Maggio VL, Patterson DG, Jr, et al. Isotope dilution-mass spectrometric quantification of specific proteins: model application with apolipoprotein A-I. Clin. Chem. 1996;42(10):1676–1682. [PubMed] [Google Scholar]
- 10.Bondar OP, Barnidge DR, Klee EW, Davis BJ, Klee GG. LC-MS/MS quantification of Zn-α2 glycoprotein: a potential serum biomarker for prostate cancer. Clin. Chem. 2007;53(4):673–678. doi: 10.1373/clinchem.2006.079681. [DOI] [PubMed] [Google Scholar]
- 11. Chen Y, Snyder MR, Zhu Y, et al. Simultaneous phenotyping and quantification of alpha-1-antrypsin by liquid chromatography-tandem mass spectrometry. Clin. Chem. 2011;57(8):1161–1168. doi: 10.1373/clinchem.2011.163006.▪ Elegant paper that describes a method to genotype and phenotype patients in a single assay.
- 12.Fierens C, StockL D, Baetens D, De Leenheer AP, Thienpont LM. Application of a C-peptide electrospray ionization-isotope dilution liquid chromatography-tandem mass spectrometry measurement procedure for the evaluation of five C-peptide immunoassays for urine. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2003;792(2):249–259. doi: 10.1016/s1570-0232(03)00268-x. [DOI] [PubMed] [Google Scholar]
- 13.Kaiser P, Akerboom T, Ohlendorf R, Reinauer H. Liquid chromatography-isotope dilution-mass spectrometry as a new basis for the reference measurement procedure for hemoglobin Alc determination. Clin. Chem. 2010;56(5):750–754. doi: 10.1373/clinchem.2009.139477. [DOI] [PubMed] [Google Scholar]
- 14.Kippen AD, Cerini F, Vadas L, et al. Development of an isotope dilution assay for precise determination of insulin, C-peptide, and proinsulin levels in nondiabetic and Type II diabetic individuals with comparison to immunoassay/ Biol. Chem. 1997;272(19):12513–12522. doi: 10.1074/jbc.272.19.12513. [DOI] [PubMed] [Google Scholar]
- 15.Kuhn E, Wu J, Karl J, Liao H, Zolg W, Guild B. Quantification of C-reactive protein in the serum of patients with rheumatoid arthritis using multiple reaction monitoring mass spectrometry and 13C-labeled peptide standards. Proteomics. 2004;4(4):1175–1186. doi: 10.1002/pmic.200300670. [DOI] [PubMed] [Google Scholar]
- 16.Kuzyk Ma, Smith D, Yang J, et al. Multiple reaction monioring-based, multiplexed, absolute quantitation of 45 proteins in human plasma. Mol. Cell. Proteomics. 2009;8(8):1860–1877. doi: 10.1074/mcp.M800540-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mckay MJ, Sherman J, Laver MT, Baker MS, Clarke SJ, Molloy MP. The development of multiple reaction monitoring assays for liver-derived plasma proteins. Proteomics Clin. Applic. 2007;1(12):1570–1581. doi: 10.1002/prca.200700305. [DOI] [PubMed] [Google Scholar]
- 18. Seegmiller JC, Barnidge DR, Burns BE, Larson TS, Lieske JC, Kumar R. Quantification of urinary albumin by using protein cleavage and LC-MS/MS. Clin. Chem. 2009;55(6):1100–1107. doi: 10.1373/clinchem.2008.115543.▪ First paper to demonstrate the utility of stable isotope-labeled internal standard proteins in the quantification of proteins in actual clinical specimens.
- 19.Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW. Mass spectrometric quantitation of peptides and proteins using stable isotope standards and capture by antipeptide antibodies (SISCAPA) Proteome Res. 2004;3(2):235–244. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- 20.Hoofnagle AN, Becker JO, Wener MH, Heinecke JW. Quantification of thyroglobulin, a low-abundance serum protein, by immunoaffinity peptide enrichment and tandem mass spectrometry. Clin. Chem. 2008;54(11):1796–1804. doi: 10.1373/clinchem.2008.109652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn E, Addona T, Keshishian H, et al. Developing multiplexed assays for troponin I and interleukin-33 in plasma by peptide immunoaffinity enrichment and targeted mass spectrometry. Clin. Chem. 2009;55(6):1108–1117. doi: 10.1373/clinchem.2009.123935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neubert H, Gale J, Muirhead D. Online high-flow peptide immunoaffinity enrichment and nanoflow LC-MS/MS: assay development for total salivary pepsin/pepsinogen. Clin. Chem. 2010;56(9):1413–1423. doi: 10.1373/clinchem.2010.144576. [DOI] [PubMed] [Google Scholar]
- 23.Whiteaker JR, Zhang H, Zhao L, et al. Integrated pipeline for mass spectrometry-based discovery and confirmation of biomarkers demonstrated in a mouse model of breast cancer. J. Proteome Res. 2007;6(10):3962–3975. doi: 10.1021/pr070202v. [DOI] [PubMed] [Google Scholar]
- 24.Anderson Nl, Jackson A, Smith D, Hardie D, Borchers C, Pearson Tw. SISCAPA peptide enrichment on magnetic beads using an in-line bead trap device. Mol. Cell. Proteomics. 2009;8(5):995–1005. doi: 10.1074/mcp.M800446-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Addona TA, Abbatiello SE, Schilling B, et al. Multiste assessment of the precision and reproducibility of multiple reaction monkoring-based measurements of proteins in plasma. Nat. Biotechnol. 2009;27(7):633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoofnagle AN. Quantitative clinical proteomics by liquid chromatography-tandem mass spectrometry: assessing the platform. Clin. Chem. 2010;56(2):161–164. doi: 10.1373/clinchem.2009.134049. [DOI] [PubMed] [Google Scholar]
- 27.Wu SL, Amato H, Biringer R, Choudhary G, Shieh P, Hancock WS. Targeted proteomics of low-level proteins in human plasma by LC-MSn: using human growth hormone as a model system. J. Proteome Res. 2002;1(5):459–465. doi: 10.1021/pr025537l. [DOI] [PubMed] [Google Scholar]
- 28.Niederkofler EE, Kiernan UA, O’Rear J, et al. Detection of endogenous B-type natriuretic peptide at very low concentrations in patients with heart failure. Circulation. 2008;1(4):258–264. doi: 10.1161/CIRCHEARTFAILURE.108.790774. [DOI] [PubMed] [Google Scholar]
- 29.Lopez MF, Rezai T, Sarracino DA, et al. Selected reaction monitoring mass spectrometric immunoassay responsive to parathyroid hormone and related variants. Clin. Chem. 2010;56(2):281–290. doi: 10.1373/clinchem.2009.137323. [DOI] [PubMed] [Google Scholar]
- 30.Maclean B, Tomazela DM, Abbatiello SE, et al. Effect of collision energy optimization on the measurement of peptides by selected reaction monitoring (SRM) mass spectrometry. Anal. Chem. 2010;82(24):10116–10124. doi: 10.1021/ac102179j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maclean B, Tomazela DM, Shulman N, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–968. doi: 10.1093/bioinformatics/btq054.▪▪ The Skyline software platform has changed the way scientists develop novel targeted proteomics assays. The software is described in this paper first, which also describes the context in which it might be used. Consequently, all novel assays should start with the workflow described in this paper.
- 32. Abbatiello SE, Mani Dr, Keshishian H, Carr SA. Automated detection of inaccurate and imprecise transitions in peptide quantification by multiple reaction monitoring mass spectrometry. Clin. Chem. 2010;56(2):291–305. doi: 10.1373/clinchem.2009.138420.▪ Describes a method to detect isobaric interferences in putative multiple reaction monitoring transitions.
- 33.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol. Syst. Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whiteaker JR, Zhao L, Anderson L, Paulovich AG. An automated and multiplexed method for high throughput peptide immunoaffinity enrichment and multiple reaction monitoring mass spectrometry-based quantification of protein biomarkers. Mol. Cell. Proteomics. 2010;9(1):184–196. doi: 10.1074/mcp.M900254-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deutsch EW, Eng JK, Zhang H, et al. Human plasma peptide atlas. Proteomics. 2005;5(13):3497–3500. doi: 10.1002/pmic.200500160. [DOI] [PubMed] [Google Scholar]
- 36.Ducret A, Van Oostveen I, Eng JK, Yates JR, Aebersold R. High throughput protein characterization by automated reverse-phase chromatography-electrospray tandem mass spectrometry. Protein Sci. 1998;7(3):706–719. doi: 10.1002/pro.5560070320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diament BJ, Noble WS. Faster SEQUEST searching for peptide identification from tandem mass spectra. J. Proteome Res. 2011;10(9):3871–3879. doi: 10.1021/pr101196n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falkner J, Andrews P. Fast tandem mass spectra-based protein identification regardless of the number of spectra or potential modifications examined. Bioinformatics. 2005;21(10):2177–2184. doi: 10.1093/bioinformatics/bti362. [DOI] [PubMed] [Google Scholar]
- 39.Schoenherr RM, Zhao L, Whiteaker JR, et al. Automated screening of monoclonal antibodies for SISCAPA assays using a magnetic bead processor and liquid chromatography–selected reaction monkoring-mass spectrometry. J. munol. Methods. 2010;353(1–2):49–61. doi: 10.1016/j.jim.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Picotti P, Lam H, Campbell D, et al. A database of mass spectrometric assays for the yeast proteome. Nat. Methods. 2008;5(11):913–914. doi: 10.1038/nmeth1108-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schoenherr RM, Zhao L, Whiteaker, et al. Automated screening of monoclonal antibodies for SISCAPA assays using a magnetic bead processor and liquid chromatography-selected reaction monkoring-mass spectrometry. J. munol. Methods. 2010;353(1–2):49–61. doi: 10.1016/j.jim.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whiteaker JR, Zhao L, Abbatiello SE, et al. Evaluation of large scale quantitative proteomic assay development using peptide affinity-based mass spectrometry. Mol. Cell. Proteomics. 2011;10(4) doi: 10.1074/mcp.M110.005645. M110 005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spieker-Polet H, Sethupathi P, Yam PC, Knight KL. Rabbit monoclonal antibodies: generating a fusion partner to produce rabbk-rabbk hybridomas. Proc. Natl Acad. Sci. USA. 1995;92(20):9348–9352. doi: 10.1073/pnas.92.20.9348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whiteaker JR, Zhao L, Zhang HY, et al. Antibody-based enrichment of peptides on magnetic beads for mass-spectrometry-based quantification of serum biomarkers. Anal. Biochem. 2007;362(1):44–54. doi: 10.1016/j.ab.2006.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahn YH, Lee JY, Kim YS, Ko JH, Yoo JS. Quantitative analysis of an aberrant glycoform of TIMPl from colon cancer serum by L-PHA-enrichment and SISCAPA with MRM mass spectrometry. J. Proteome Res. 2009;8(9):4216–4224. doi: 10.1021/pr900269s. [DOI] [PubMed] [Google Scholar]
- 46.Annesley TM. Ionsuppression in mass spectrometry. Clin. Chem. 2003;49(7):1041–1044. doi: 10.1373/49.7.1041. [DOI] [PubMed] [Google Scholar]
- 47.Borchers CH, Parker CE. Improving the biomarker pipeline. Clin. Chem. 2010;56(12):1786–1788. doi: 10.1373/clinchem.2010.155705. [DOI] [PubMed] [Google Scholar]
- 48.Beynon RJ, Doherty MK, Pratt JM, Gaskell SJ. Multiplexed absolute quantification in proteomics using artificial QCAT proteins of concatenated signature peptides. Nat. Methods. 2005;2(8):587–589. doi: 10.1038/nmeth774. [DOI] [PubMed] [Google Scholar]
- 49.Brun V, Dupuis A, Adrait A, et al. Isotope-labeled protein standards: toward absolute quantitative proteomics. Mol. Cell. Proteomics. 2007;6(12):2139–2149. doi: 10.1074/mcp.M700163-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.Kelly RT, Tolmachev AV, Page JS, Tang K, Smith RD. The ion funnel: theory, implementations, and applications. Mass Spectrom. Rev. 2010;29(2):294–312. doi: 10.1002/mas.20232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adkins JN, Varnum Sm, Auberry KJ, et al. Toward a human blood serum proteome: analysis by multidimensional separation coupled with mass spectrometry. Mol. Cell. Proteomics. 2002;1(12):947–955. doi: 10.1074/mcp.m200066-mcp200. [DOI] [PubMed] [Google Scholar]
- 52.Thomas A, Schanzer W, Delahaut P, Thevis M. Immunoaffinity purification of peptide hormones prior to liquid chromatography–mass spectrometry in doping controls. Methods. 2011 doi: 10.1016/j.ymeth.2011.08.009. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 53.Oran PE, Jarvis JW, Borges CR, Sherma ND, Nelson RW. Mass spectro metric immunoassay of intact insulin and related variants for population proteomics studies. Proteomics Clin. Appl. 2011;5(7–8):454–459. doi: 10.1002/prca.201000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Niederkofler EE, Kiernan UA, O’rear J, et al. Detection of endogenous B-type natriuretic peptide at very low concentrations in patients with heart failure. Circ. Heart Fail. 2008;1(4):258–264. doi: 10.1161/CIRCHEARTFAILURE.108.790774. [DOI] [PubMed] [Google Scholar]
- 55.Nelson RW, Nedelkov D, Tubbs KA, Kiernan UA. Quantitative mass spectrometric immunoassay of insulin like growth factor 1. J. Proteome Res. 2004;3(4):851–855. doi: 10.1021/pr0499388. [DOI] [PubMed] [Google Scholar]
- 56.Nedelkov D, Kiernan UA, Niederkofler EE, Tubbs KA, Nelson RW. Investigating diversity in human plasma proteins. Proc. Natl. Acad. Sci. USA. 2005;102(31):10852–10857. doi: 10.1073/pnas.0500426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Borges CR, Oran PE, Buddi S, et al. Building multidimensional biomarker views of Type 2 diabetes on the basis of protein microheterogeneky. Clin. Chem. 2011;57(5):719–728. doi: 10.1373/clinchem.2010.156976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pierce CL, Williams TL, Moura H, et al. Quantification of immunoreactive viral influenza proteins by immunoaffinity capture and isotope-dilution liquid chromatography-tandem mass spectrometry. Anal. Chem. 2011;83(12):4729–4737. doi: 10.1021/ac2006526. [DOI] [PubMed] [Google Scholar]
- 59.Fortin T, Salvador A, Charrier JP, et al. Clinical quantitation of prostate-specific antigen biomarker in the low nanogram/ milliliter range by conventional bore liquid chromatography-tandem mass spectrometry (multiple reaction monitoring) coupling and correlation with ELISA tests. Mol. Cell. Proteomics. 2009;8(5):1006–1015. doi: 10.1074/mcp.M800238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kiernan UA, Addobbati R, Nedelkov D, Nelson RW. Quantitative multiplexed C-reactive protein mass spectrometric immunoassay. J. Proteome Res. 2006;5(7):1682–1687. doi: 10.1021/pr0601133. [DOI] [PubMed] [Google Scholar]
- 61.Zhao Y, Widen SG, Jamaluddin M, et al. Quantification of activated NF-kappaB/RelA complexes using ssDNA aptamer affinity-stable isotope dilution-selected reaction monitoring–mass spectrometry. Mol. Cell. Proteomics. 2011;10(6) doi: 10.1074/mcp.M111.008771. Mill 008771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shushan B. A review of clinical diagnostic applications of liquid chromatography-tandem mass spectrometry. Mass Spectrom. Rev. 2010;29(6):930–944. doi: 10.1002/mas.20295. [DOI] [PubMed] [Google Scholar]
Website
- 101.Office of cancer clinical proteomics research antibody portal. http://antibodies.cancer.gov.