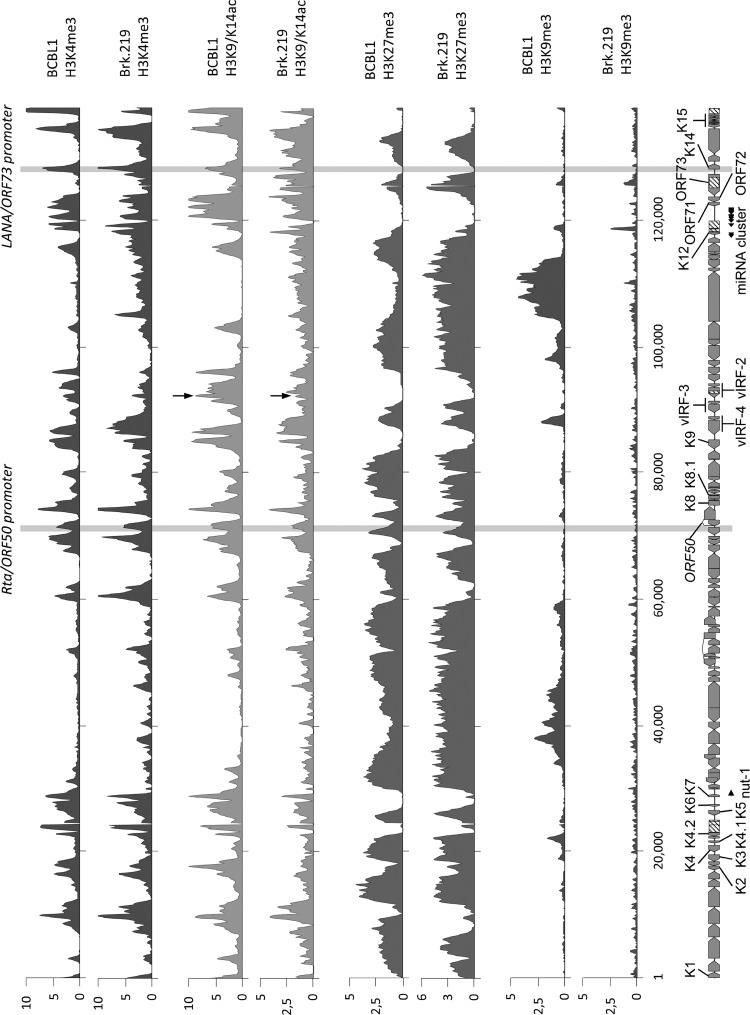
Fig 2.
Comparison of global histone modification patterns of KSHV genomes in BrK.219 and PEL cells. Global patterns of histone H3 trimethylated at lysine 4 (H3K4-me3), lysine 9 (H3K9-me3), or lysine 27 (H3K27-me3) or acetylated at lysine 9 and/or lysine 14 (H3K27-me3 and H3K9/14-ac). Modification patterns in BrK.219 cells were analyzed by ChIP-on-chip analysis using high-resolution KSHV tiling microarrays. Values shown on the y axis represent relative enrichment of normalized signals from the immunoprecipitated over input material. Histone modification patterns in the PEL line BCBL-1 (47) are shown for comparison. Regions highlighted in gray indicate the position of the ORF50/Rta and the ORF73/LANA promoters, both of which carry activating H3K4-me3 and H3K9/14-ac marks. In addition, an H3K19/14-ac peak, located upstream of the vIRF-3 coding region and marked by arrows, is more prominent in BCBL-1 compared to BrK.219 cells, which may reflect the lower basal expression levels of vIRF-3 during latency in BrK.219 cells. The ORF50 promoter additionally acquires repressive H3K27-me3 marks and thus exhibits the hallmarks of bivalent chromatin, which is transcriptionally silent, but “poised” for reactivation (47).