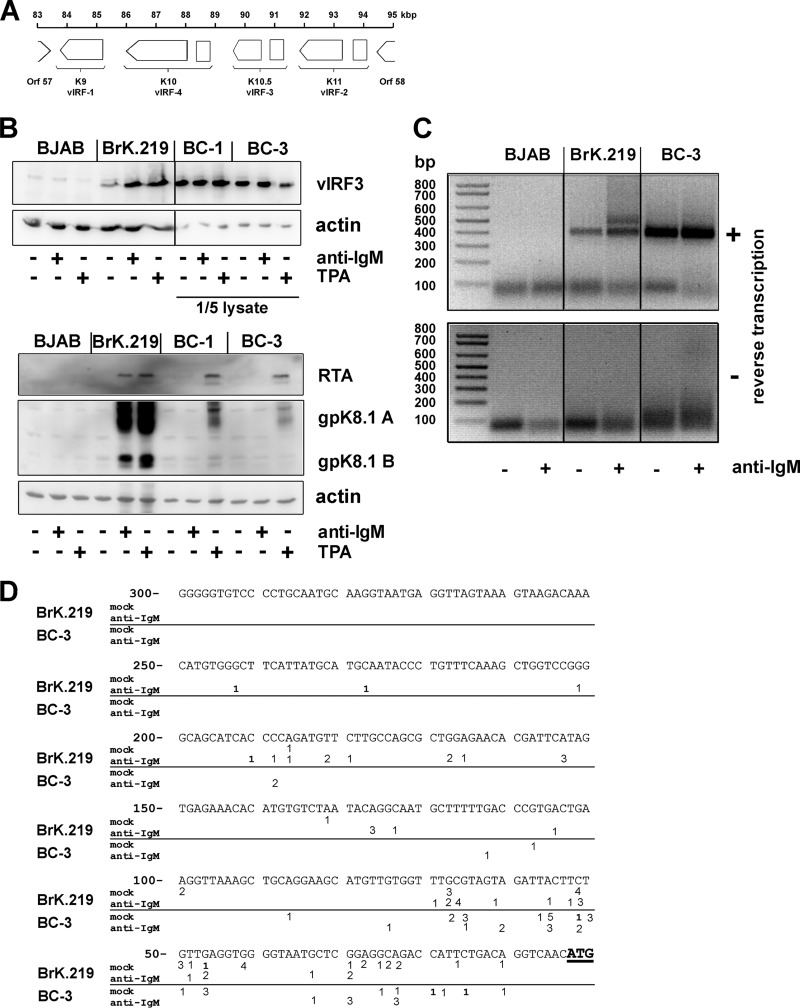
Fig 5.
vIRF-3 is an inducible gene with two 5′ start sites in BrK.219 cells. (A) Gene arrangement in the vIRF region shown between ORFs 57 and 58. (B) BJAB cells, their rKSHV.219-infected counterparts (BrK.219), and PEL cell lines (BC-3 and BC-1) were treated with either 2.5 μg of anti-IgM/ml or 25 ng of TPA/ml for 3 days or left untreated as a control. Viral protein expression was investigated by immunoblotting. For analysis of vIRF-3 expression in PEL cells, only 1/5 of the cell lysates were used. (C) 5′-RACE of vIRF-3 transcripts. BrK.219 cells, uninfected parental BJAB cells, and the PEL cell line BC-3 were treated for 3 days with anti-IgM or left untreated. Reverse transcription and PCR was performed with total RNAs. Agarose gels show the products of the 5′-RACE, which was also performed on RNA samples without reverse transcription to control for the potential amplification of contaminating DNA. (D) Location of 5′ start sites of vIRF-3 mRNAs determined by 5′-RACE. The 5′-RACE products as shown in panel C of two independent experiments were cloned, and multiple clones were sequenced to determine their 5′ ends. Numbers (ranging between 1 and 5) reflect the frequency and position of individual 5′ start sites. Whereas in BC-3 cells most transcripts originate within the first 100 bp upstream of the vIRF-3 ATG, in anti-IgM-stimulated BrK.219 cells, additional peaks of initiation are located up to further 150 bp upstream at position −101 to position −250.