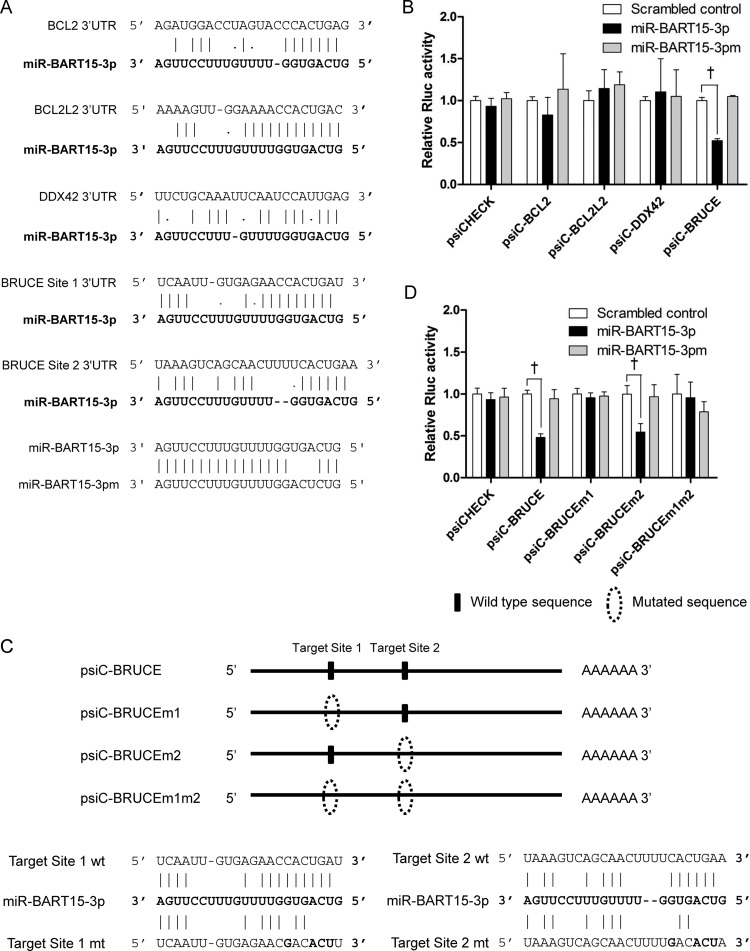
Fig 4.
Target search for miR-BART15-3p. (A) Seed-matched regions of the putative target genes for miR-BART15-3p are shown. (B) To test whether the putative target genes are regulated by miR-BART15-3p, the 3′ UTR fragment of each gene was cloned into a luciferase reporter (psiCHECK-2) vector. HEK293T cells were cotransfected with the miR-BART15-3p mimic and the appropriate 3′ UTR reporter plasmid. To confirm the sequence-specific function of miR-BART15-3p, miR-BART15-3pm (the sequence is shown at the bottom of panel A), which has substituted nucleotides at ∼4 to 6 sites of miR-BART15-3p, was also used. Luciferase activity was analyzed 48 h after transfection (n = 3). (C) Illustration showing the location of the possible seed-matched sites for miR-BART15-3p on the BRUCE 3′ UTR and the sites changed to produce mutated forms of psiC-BRUCE. (D) HEK293T cells were cotransfected with miR-BART15-3p and the indicated psiC-BRUCE vector containing the wild-type or mutated 3′ UTR of BRUCE. Luciferase activity was normalized using firefly luciferase activity. Error bars indicate SD (n = 3). †, P < 0.01.